Maciej Śliwowski
Deep learning for ECoG brain-computer interface: end-to-end vs. hand-crafted features
Oct 05, 2022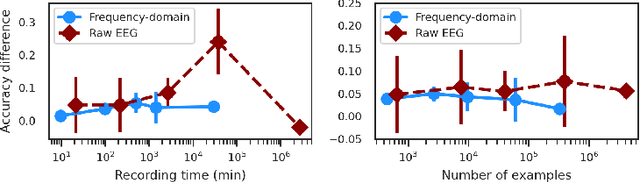
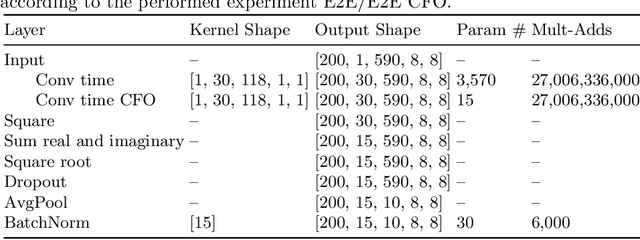

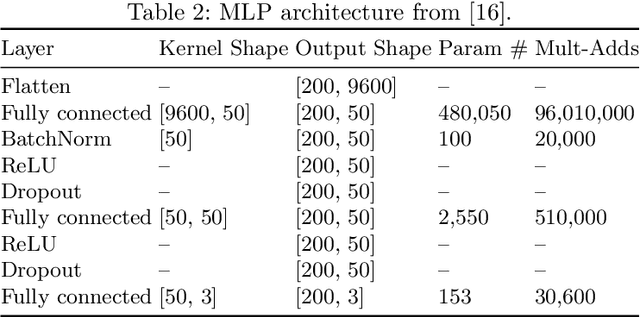
Abstract:In brain signal processing, deep learning (DL) models have become commonly used. However, the performance gain from using end-to-end DL models compared to conventional ML approaches is usually significant but moderate, typically at the cost of increased computational load and deteriorated explainability. The core idea behind deep learning approaches is scaling the performance with bigger datasets. However, brain signals are temporal data with a low signal-to-noise ratio, uncertain labels, and nonstationary data in time. Those factors may influence the training process and slow down the models' performance improvement. These factors' influence may differ for end-to-end DL model and one using hand-crafted features. As not studied before, this paper compares models that use raw ECoG signal and time-frequency features for BCI motor imagery decoding. We investigate whether the current dataset size is a stronger limitation for any models. Finally, obtained filters were compared to identify differences between hand-crafted features and optimized with backpropagation. To compare the effectiveness of both strategies, we used a multilayer perceptron and a mix of convolutional and LSTM layers that were already proved effective in this task. The analysis was performed on the long-term clinical trial database (almost 600 minutes of recordings) of a tetraplegic patient executing motor imagery tasks for 3D hand translation. For a given dataset, the results showed that end-to-end training might not be significantly better than the hand-crafted features-based model. The performance gap is reduced with bigger datasets, but considering the increased computational load, end-to-end training may not be profitable for this application.
Impact of dataset size and long-term ECoG-based BCI usage on deep learning decoders performance
Sep 08, 2022

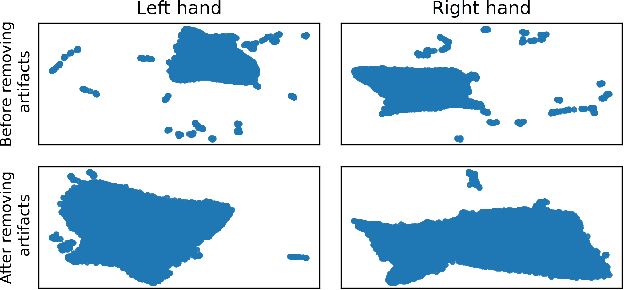
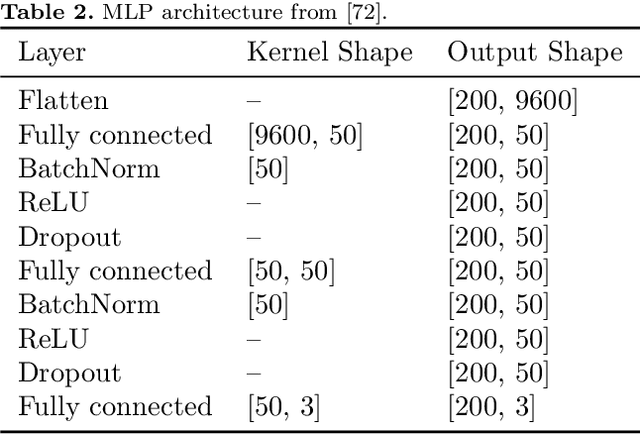
Abstract:In brain-computer interfaces (BCI) research, recording data is time-consuming and expensive, which limits access to big datasets. This may influence the BCI system performance as machine learning methods depend strongly on the training dataset size. Important questions arise: taking into account neuronal signal characteristics (e.g., non-stationarity), can we achieve higher decoding performance with more data to train decoders? What is the perspective for further improvement with time in the case of long-term BCI studies? In this study, we investigated the impact of long-term recordings on motor imagery decoding from two main perspectives: model requirements regarding dataset size and potential for patient adaptation. We evaluated the multilinear model and two deep learning (DL) models on a long-term BCI and Tetraplegia NCT02550522 clinical trial dataset containing 43 sessions of ECoG recordings performed with a tetraplegic patient. In the experiment, a participant executed 3D virtual hand translation using motor imagery patterns. We designed multiple computational experiments in which training datasets were increased or translated to investigate the relationship between models' performance and different factors influencing recordings. Our analysis showed that adding more data to the training dataset may not instantly increase performance for datasets already containing 40 minutes of the signal. DL decoders showed similar requirements regarding the dataset size compared to the multilinear model while demonstrating higher decoding performance. Moreover, high decoding performance was obtained with relatively small datasets recorded later in the experiment, suggesting motor imagery patterns improvement and patient adaptation. Finally, we proposed UMAP embeddings and local intrinsic dimensionality as a way to visualize the data and potentially evaluate data quality.
2021 BEETL Competition: Advancing Transfer Learning for Subject Independence & Heterogenous EEG Data Sets
Feb 14, 2022
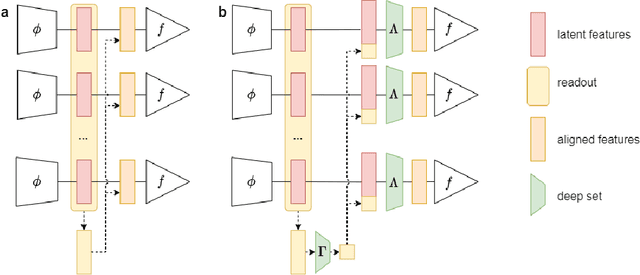
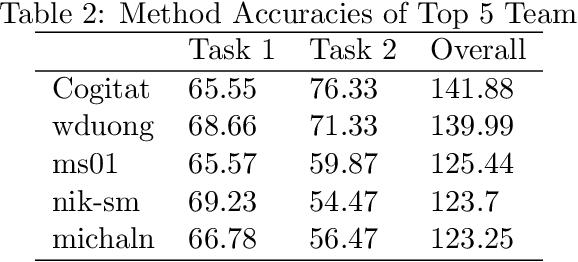
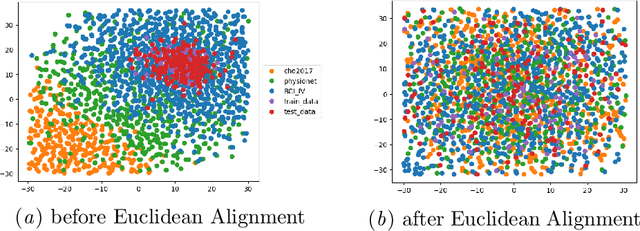
Abstract:Transfer learning and meta-learning offer some of the most promising avenues to unlock the scalability of healthcare and consumer technologies driven by biosignal data. This is because current methods cannot generalise well across human subjects' data and handle learning from different heterogeneously collected data sets, thus limiting the scale of training data. On the other side, developments in transfer learning would benefit significantly from a real-world benchmark with immediate practical application. Therefore, we pick electroencephalography (EEG) as an exemplar for what makes biosignal machine learning hard. We design two transfer learning challenges around diagnostics and Brain-Computer-Interfacing (BCI), that have to be solved in the face of low signal-to-noise ratios, major variability among subjects, differences in the data recording sessions and techniques, and even between the specific BCI tasks recorded in the dataset. Task 1 is centred on the field of medical diagnostics, addressing automatic sleep stage annotation across subjects. Task 2 is centred on Brain-Computer Interfacing (BCI), addressing motor imagery decoding across both subjects and data sets. The BEETL competition with its over 30 competing teams and its 3 winning entries brought attention to the potential of deep transfer learning and combinations of set theory and conventional machine learning techniques to overcome the challenges. The results set a new state-of-the-art for the real-world BEETL benchmark.
Decoding ECoG signal into 3D hand translation using deep learning
Oct 05, 2021


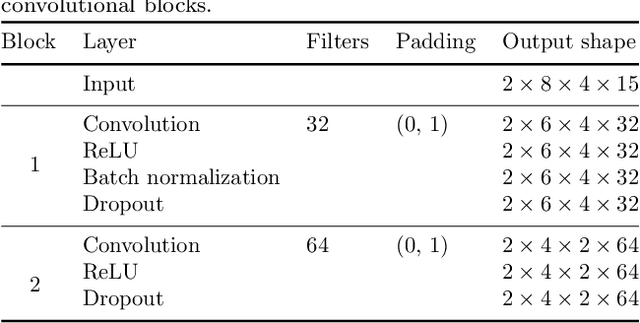
Abstract:Motor brain-computer interfaces (BCIs) are a promising technology that may enable motor-impaired people to interact with their environment. Designing real-time and accurate BCI is crucial to make such devices useful, safe, and easy to use by patients in a real-life environment. Electrocorticography (ECoG)-based BCIs emerge as a good compromise between invasiveness of the recording device and good spatial and temporal resolution of the recorded signal. However, most ECoG signal decoders used to predict continuous hand movements are linear models. These models have a limited representational capacity and may fail to capture the relationship between ECoG signal and continuous hand movements. Deep learning (DL) models, which are state-of-the-art in many problems, could be a solution to better capture this relationship. In this study, we tested several DL-based architectures to predict imagined 3D continuous hand translation using time-frequency features extracted from ECoG signals. The dataset used in the analysis is a part of a long-term clinical trial (ClinicalTrials.gov identifier: NCT02550522) and was acquired during a closed-loop experiment with a tetraplegic subject. The proposed architectures include multilayer perceptron (MLP), convolutional neural networks (CNN), and long short-term memory networks (LSTM). The accuracy of the DL-based and multilinear models was compared offline using cosine similarity. Our results show that CNN-based architectures outperform the current state-of-the-art multilinear model. The best architecture exploited the spatial correlation between neighboring electrodes with CNN and benefited from the sequential character of the desired hand trajectory by using LSTMs. Overall, DL increased the average cosine similarity, compared to the multilinear model, by up to 60%, from 0.189 to 0.302 and from 0.157 to 0.249 for the left and right hand, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge