Siegfried Ludwig
EEG-D3: A Solution to the Hidden Overfitting Problem of Deep Learning Models
Dec 15, 2025Abstract:Deep learning for decoding EEG signals has gained traction, with many claims to state-of-the-art accuracy. However, despite the convincing benchmark performance, successful translation to real applications is limited. The frequent disconnect between performance on controlled BCI benchmarks and its lack of generalisation to practical settings indicates hidden overfitting problems. We introduce Disentangled Decoding Decomposition (D3), a weakly supervised method for training deep learning models across EEG datasets. By predicting the place in the respective trial sequence from which the input window was sampled, EEG-D3 separates latent components of brain activity, akin to non-linear ICA. We utilise a novel model architecture with fully independent sub-networks for strict interpretability. We outline a feature interpretation paradigm to contrast the component activation profiles on different datasets and inspect the associated temporal and spatial filters. The proposed method reliably separates latent components of brain activity on motor imagery data. Training downstream classifiers on an appropriate subset of these components prevents hidden overfitting caused by task-correlated artefacts, which severely affects end-to-end classifiers. We further exploit the linearly separable latent space for effective few-shot learning on sleep stage classification. The ability to distinguish genuine components of brain activity from spurious features results in models that avoid the hidden overfitting problem and generalise well to real-world applications, while requiring only minimal labelled data. With interest to the neuroscience community, the proposed method gives researchers a tool to separate individual brain processes and potentially even uncover heretofore unknown dynamics.
Stochastic Resonance Improves the Detection of Low Contrast Images in Deep Learning Models
Feb 20, 2025Abstract:Stochastic resonance describes the utility of noise in improving the detectability of weak signals in certain types of systems. It has been observed widely in natural and engineered settings, but its utility in image classification with rate-based neural networks has not been studied extensively. In this analysis a simple LSTM recurrent neural network is trained for digit recognition and classification. During the test phase, image contrast is reduced to a point where the model fails to recognize the presence of a stimulus. Controlled noise is added to partially recover classification performance. The results indicate the presence of stochastic resonance in rate-based recurrent neural networks.
Latent Alignment with Deep Set EEG Decoders
Nov 29, 2023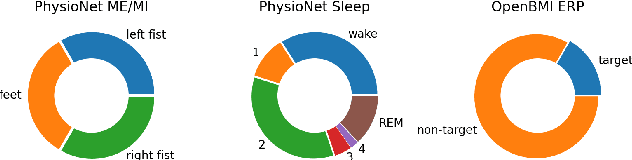
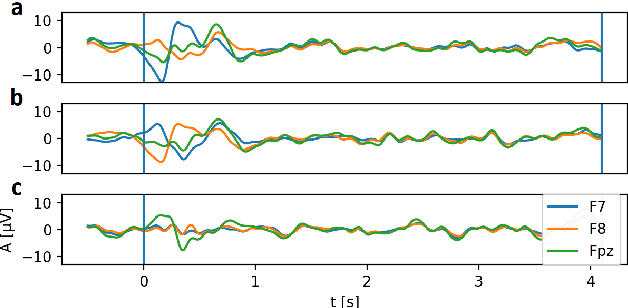
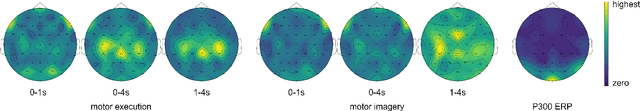
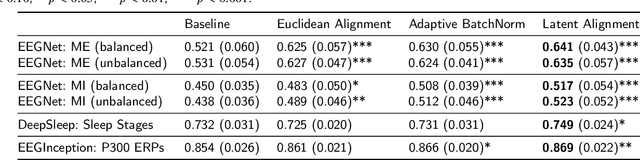
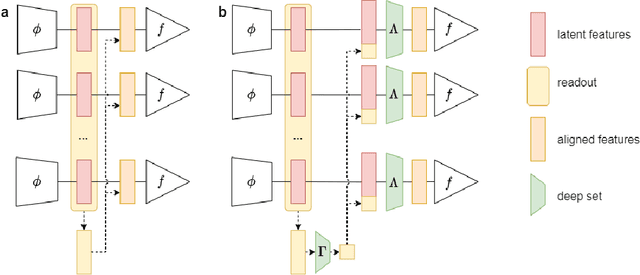
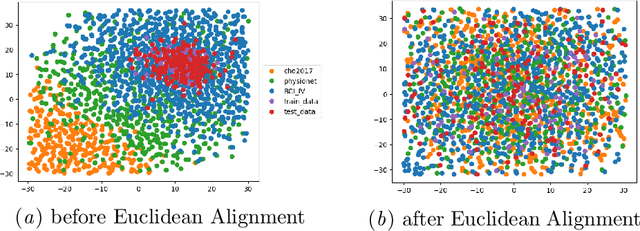
Abstract:The variability in EEG signals between different individuals poses a significant challenge when implementing brain-computer interfaces (BCI). Commonly proposed solutions to this problem include deep learning models, due to their increased capacity and generalization, as well as explicit domain adaptation techniques. Here, we introduce the Latent Alignment method that won the Benchmarks for EEG Transfer Learning (BEETL) competition and present its formulation as a deep set applied on the set of trials from a given subject. Its performance is compared to recent statistical domain adaptation techniques under various conditions. The experimental paradigms include motor imagery (MI), oddball event-related potentials (ERP) and sleep stage classification, where different well-established deep learning models are applied on each task. Our experimental results show that performing statistical distribution alignment at later stages in a deep learning model is beneficial to the classification accuracy, yielding the highest performance for our proposed method. We further investigate practical considerations that arise in the context of using deep learning and statistical alignment for EEG decoding. In this regard, we study class-discriminative artifacts that can spuriously improve results for deep learning models, as well as the impact of class-imbalance on alignment. We delineate a trade-off relationship between increased classification accuracy when alignment is performed at later modeling stages, and susceptibility to class-imbalance in the set of trials that the statistics are computed on.
2021 BEETL Competition: Advancing Transfer Learning for Subject Independence & Heterogenous EEG Data Sets
Feb 14, 2022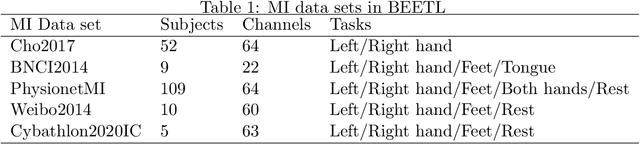
Abstract:Transfer learning and meta-learning offer some of the most promising avenues to unlock the scalability of healthcare and consumer technologies driven by biosignal data. This is because current methods cannot generalise well across human subjects' data and handle learning from different heterogeneously collected data sets, thus limiting the scale of training data. On the other side, developments in transfer learning would benefit significantly from a real-world benchmark with immediate practical application. Therefore, we pick electroencephalography (EEG) as an exemplar for what makes biosignal machine learning hard. We design two transfer learning challenges around diagnostics and Brain-Computer-Interfacing (BCI), that have to be solved in the face of low signal-to-noise ratios, major variability among subjects, differences in the data recording sessions and techniques, and even between the specific BCI tasks recorded in the dataset. Task 1 is centred on the field of medical diagnostics, addressing automatic sleep stage annotation across subjects. Task 2 is centred on Brain-Computer Interfacing (BCI), addressing motor imagery decoding across both subjects and data sets. The BEETL competition with its over 30 competing teams and its 3 winning entries brought attention to the potential of deep transfer learning and combinations of set theory and conventional machine learning techniques to overcome the challenges. The results set a new state-of-the-art for the real-world BEETL benchmark.
Team Cogitat at NeurIPS 2021: Benchmarks for EEG Transfer Learning Competition
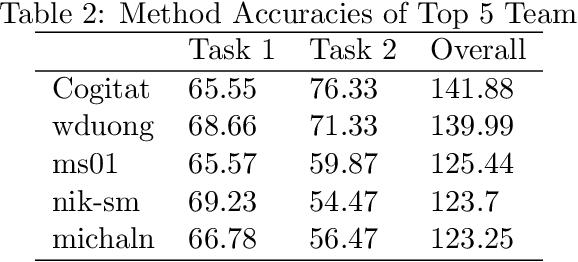
Feb 01, 2022Abstract:Building subject-independent deep learning models for EEG decoding faces the challenge of strong covariate-shift across different datasets, subjects and recording sessions. Our approach to address this difficulty is to explicitly align feature distributions at various layers of the deep learning model, using both simple statistical techniques as well as trainable methods with more representational capacity. This follows in a similar vein as covariance-based alignment methods, often used in a Riemannian manifold context. The methodology proposed herein won first place in the 2021 Benchmarks in EEG Transfer Learning (BEETL) competition, hosted at the NeurIPS conference. The first task of the competition consisted of sleep stage classification, which required the transfer of models trained on younger subjects to perform inference on multiple subjects of older age groups without personalized calibration data, requiring subject-independent models. The second task required to transfer models trained on the subjects of one or more source motor imagery datasets to perform inference on two target datasets, providing a small set of personalized calibration data for multiple test subjects.
EEGminer: Discovering Interpretable Features of Brain Activity with Learnable Filters
Oct 19, 2021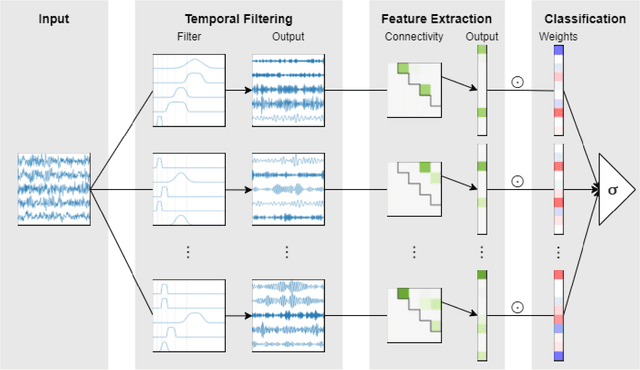
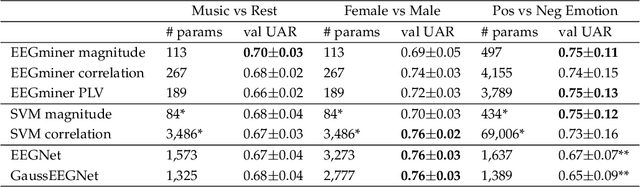
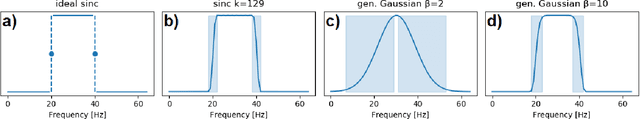
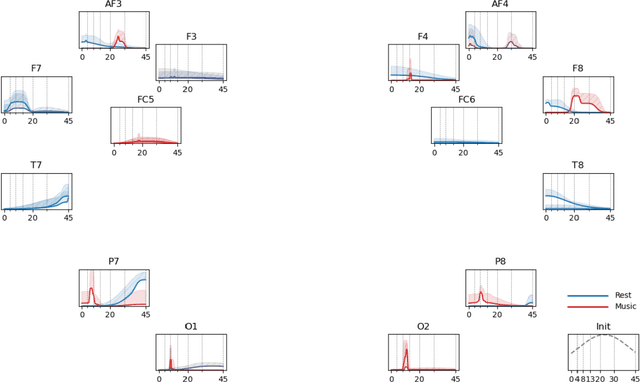
Abstract:Patterns of brain activity are associated with different brain processes and can be used to identify different brain states and make behavioral predictions. However, the relevant features are not readily apparent and accessible. To mine informative latent representations from multichannel EEG recordings, we propose a novel differentiable EEG decoding pipeline consisting of learnable filters and a pre-determined feature extraction module. Specifically, we introduce filters parameterized by generalized Gaussian functions that offer a smooth derivative for stable end-to-end model training and allow for learning interpretable features. For the feature module, we use signal magnitude and functional connectivity. We demonstrate the utility of our model towards emotion recognition from EEG signals on the SEED dataset, as well as on a new EEG dataset of unprecedented size (i.e., 763 subjects), where we identify consistent trends of music perception and related individual differences. The discovered features align with previous neuroscience studies and offer new insights, such as marked differences in the functional connectivity profile between left and right temporal areas during music listening. This agrees with the respective specialisation of the temporal lobes regarding music perception proposed in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge