Sylvain Chevallier
TAU, LISN
EEG Foundation Challenge: From Cross-Task to Cross-Subject EEG Decoding
Jun 23, 2025Abstract:Current electroencephalogram (EEG) decoding models are typically trained on small numbers of subjects performing a single task. Here, we introduce a large-scale, code-submission-based competition comprising two challenges. First, the Transfer Challenge asks participants to build and test a model that can zero-shot decode new tasks and new subjects from their EEG data. Second, the Psychopathology factor prediction Challenge asks participants to infer subject measures of mental health from EEG data. For this, we use an unprecedented, multi-terabyte dataset of high-density EEG signals (128 channels) recorded from over 3,000 child to young adult subjects engaged in multiple active and passive tasks. We provide several tunable neural network baselines for each of these two challenges, including a simple network and demographic-based regression models. Developing models that generalise across tasks and individuals will pave the way for ML network architectures capable of adapting to EEG data collected from diverse tasks and individuals. Similarly, predicting mental health-relevant personality trait values from EEG might identify objective biomarkers useful for clinical diagnosis and design of personalised treatment for psychological conditions. Ultimately, the advances spurred by this challenge could contribute to the development of computational psychiatry and useful neurotechnology, and contribute to breakthroughs in both fundamental neuroscience and applied clinical research.
* Approved at Neurips Competition track. webpage: https://eeg2025.github.io/
A probabilistic view on Riemannian machine learning models for SPD matrices
May 05, 2025Abstract:The goal of this paper is to show how different machine learning tools on the Riemannian manifold $\mathcal{P}_d$ of Symmetric Positive Definite (SPD) matrices can be united under a probabilistic framework. For this, we will need several Gaussian distributions defined on $\mathcal{P}_d$. We will show how popular classifiers on $\mathcal{P}_d$ can be reinterpreted as Bayes Classifiers using these Gaussian distributions. These distributions will also be used for outlier detection and dimension reduction. By showing that those distributions are pervasive in the tools used on $\mathcal{P}_d$, we allow for other machine learning tools to be extended to $\mathcal{P}_d$.
Wrapped Gaussian on the manifold of Symmetric Positive Definite Matrices
Feb 03, 2025Abstract:Circular and non-flat data distributions are prevalent across diverse domains of data science, yet their specific geometric structures often remain underutilized in machine learning frameworks. A principled approach to accounting for the underlying geometry of such data is pivotal, particularly when extending statistical models, like the pervasive Gaussian distribution. In this work, we tackle those issue by focusing on the manifold of symmetric positive definite matrices, a key focus in information geometry. We introduced a non-isotropic wrapped Gaussian by leveraging the exponential map, we derive theoretical properties of this distribution and propose a maximum likelihood framework for parameter estimation. Furthermore, we reinterpret established classifiers on SPD through a probabilistic lens and introduce new classifiers based on the wrapped Gaussian model. Experiments on synthetic and real-world datasets demonstrate the robustness and flexibility of this geometry-aware distribution, underscoring its potential to advance manifold-based data analysis. This work lays the groundwork for extending classical machine learning and statistical methods to more complex and structured data.
Geodesic Optimization for Predictive Shift Adaptation on EEG data
Jul 04, 2024



Abstract:Electroencephalography (EEG) data is often collected from diverse contexts involving different populations and EEG devices. This variability can induce distribution shifts in the data $X$ and in the biomedical variables of interest $y$, thus limiting the application of supervised machine learning (ML) algorithms. While domain adaptation (DA) methods have been developed to mitigate the impact of these shifts, such methods struggle when distribution shifts occur simultaneously in $X$ and $y$. As state-of-the-art ML models for EEG represent the data by spatial covariance matrices, which lie on the Riemannian manifold of Symmetric Positive Definite (SPD) matrices, it is appealing to study DA techniques operating on the SPD manifold. This paper proposes a novel method termed Geodesic Optimization for Predictive Shift Adaptation (GOPSA) to address test-time multi-source DA for situations in which source domains have distinct $y$ distributions. GOPSA exploits the geodesic structure of the Riemannian manifold to jointly learn a domain-specific re-centering operator representing site-specific intercepts and the regression model. We performed empirical benchmarks on the cross-site generalization of age-prediction models with resting-state EEG data from a large multi-national dataset (HarMNqEEG), which included $14$ recording sites and more than $1500$ human participants. Compared to state-of-the-art methods, our results showed that GOPSA achieved significantly higher performance on three regression metrics ($R^2$, MAE, and Spearman's $\rho$) for several source-target site combinations, highlighting its effectiveness in tackling multi-source DA with predictive shifts in EEG data analysis. Our method has the potential to combine the advantages of mixed-effects modeling with machine learning for biomedical applications of EEG, such as multicenter clinical trials.
Growing Tiny Networks: Spotting Expressivity Bottlenecks and Fixing Them Optimally
May 30, 2024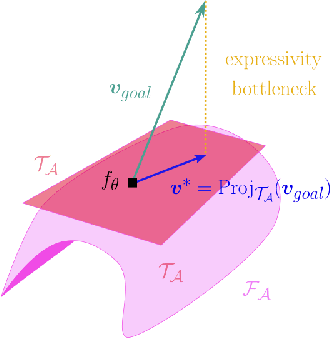
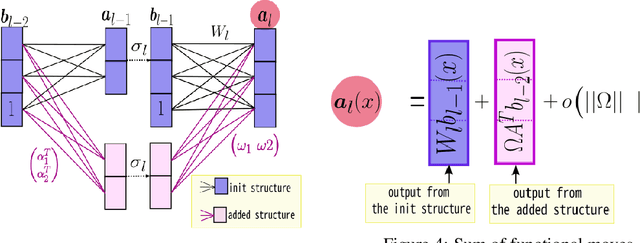
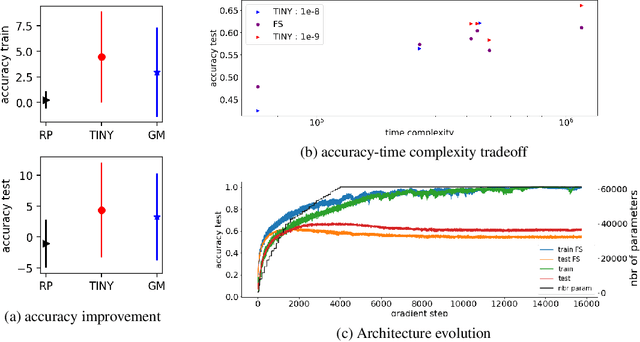
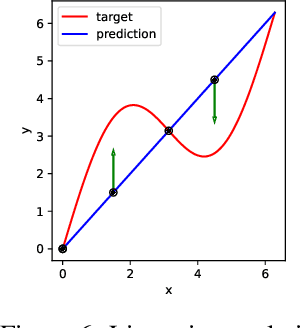
Abstract:Machine learning tasks are generally formulated as optimization problems, where one searches for an optimal function within a certain functional space. In practice, parameterized functional spaces are considered, in order to be able to perform gradient descent. Typically, a neural network architecture is chosen and fixed, and its parameters (connection weights) are optimized, yielding an architecture-dependent result. This way of proceeding however forces the evolution of the function during training to lie within the realm of what is expressible with the chosen architecture, and prevents any optimization across architectures. Costly architectural hyper-parameter optimization is often performed to compensate for this. Instead, we propose to adapt the architecture on the fly during training. We show that the information about desirable architectural changes, due to expressivity bottlenecks when attempting to follow the functional gradient, can be extracted from %the backpropagation. To do this, we propose a mathematical definition of expressivity bottlenecks, which enables us to detect, quantify and solve them while training, by adding suitable neurons when and where needed. Thus, while the standard approach requires large networks, in terms of number of neurons per layer, for expressivity and optimization reasons, we are able to start with very small neural networks and let them grow appropriately. As a proof of concept, we show results~on the CIFAR dataset, matching large neural network accuracy, with competitive training time, while removing the need for standard architectural hyper-parameter search.
Combining Euclidean Alignment and Data Augmentation for BCI decoding
May 23, 2024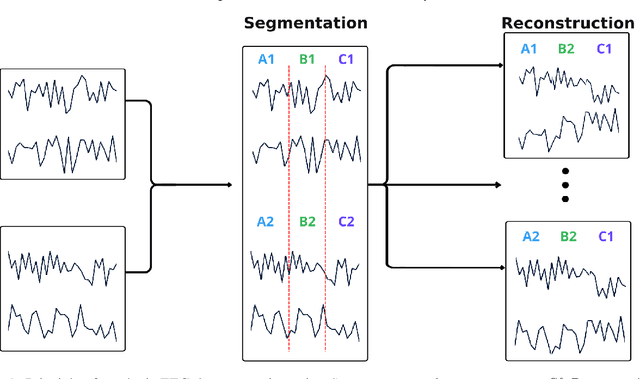
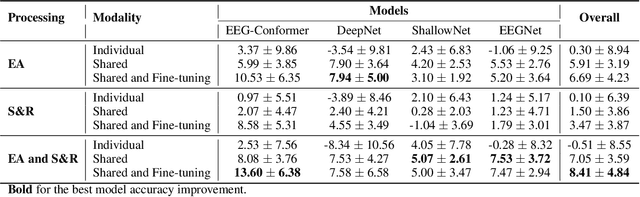
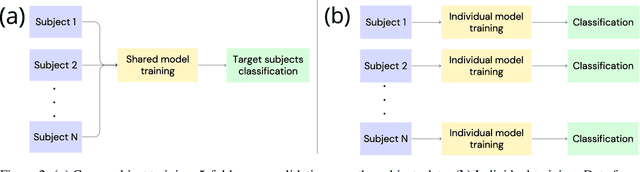
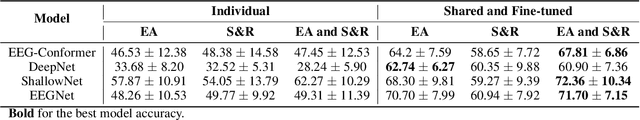
Abstract:Automated classification of electroencephalogram (EEG) signals is complex due to their high dimensionality, non-stationarity, low signal-to-noise ratio, and variability between subjects. Deep neural networks (DNNs) have shown promising results for EEG classification, but the above challenges hinder their performance. Euclidean Alignment (EA) and Data Augmentation (DA) are two promising techniques for improving DNN training by permitting the use of data from multiple subjects, increasing the data, and regularizing the available data. In this paper, we perform a detailed evaluation of the combined use of EA and DA with DNNs for EEG decoding. We trained individual models and shared models with data from multiple subjects and showed that combining EA and DA generates synergies that improve the accuracy of most models and datasets. Also, the shared models combined with fine-tuning benefited the most, with an overall increase of 8.41\% in classification accuracy.
Geometric Neural Network based on Phase Space for BCI decoding
Mar 08, 2024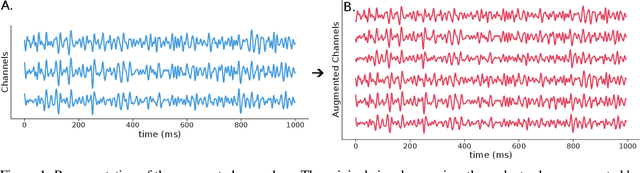
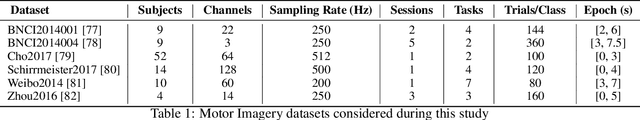
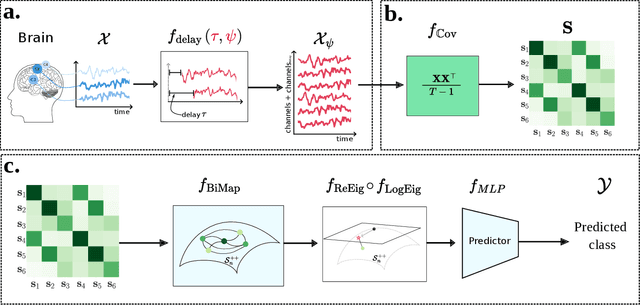
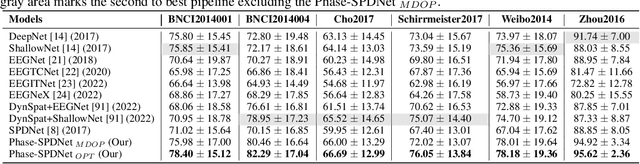
Abstract:The integration of Deep Learning (DL) algorithms on brain signal analysis is still in its nascent stages compared to their success in fields like Computer Vision, especially in Brain-Computer Interface (BCI), where the brain activity is decoded to control external devices without requiring muscle control. Electroencephalography (EEG) is a widely adopted choice for designing BCI systems due to its non-invasive and cost-effective nature and excellent temporal resolution. Still, it comes at the expense of limited training data, poor signal-to-noise, and a large variability across and within-subject recordings. Finally, setting up a BCI system with many electrodes takes a long time, hindering the widespread adoption of reliable DL architectures in BCIs outside research laboratories. To improve adoption, we need to improve user comfort using, for instance, reliable algorithms that operate with few electrodes. \textbf{Approach:} Our research aims to develop a DL algorithm that delivers effective results with a limited number of electrodes. Taking advantage of the Augmented Covariance Method with SPDNet, we propose the SPDNet$_{\psi}$ architecture and analyze its performance and computational impact, as well as the interpretability of the results. The evaluation is conducted on 5-fold cross-validation, using only three electrodes positioned above the Motor Cortex. The methodology was tested on nearly 100 subjects from several open-source datasets using the Mother Of All BCI Benchmark (MOABB) framework. \textbf{Main results:} The results of our SPDNet$_{\psi}$ demonstrate that the augmented approach combined with the SPDNet significantly outperforms all the current state-of-the-art DL architecture in MI decoding. \textbf{Significance:} This new architecture is explainable, with a low number of trainable parameters and a reduced carbon footprint.
A Systematic Evaluation of Euclidean Alignment with Deep Learning for EEG Decoding
Jan 30, 2024Abstract:Electroencephalography (EEG) signals are frequently used for various Brain-Computer Interface (BCI) tasks. While Deep Learning (DL) techniques have shown promising results, they are hindered by the substantial data requirements. By leveraging data from multiple subjects, transfer learning enables more effective training of DL models. A technique that is gaining popularity is Euclidean Alignment (EA) due to its ease of use, low computational complexity, and compatibility with Deep Learning models. However, few studies evaluate its impact on the training performance of shared and individual DL models. In this work, we systematically evaluate the effect of EA combined with DL for decoding BCI signals. We used EA to train shared models with data from multiple subjects and evaluated its transferability to new subjects. Our experimental results show that it improves decoding in the target subject by 4.33% and decreases convergence time by more than 70%. We also trained individual models for each subject to use as a majority-voting ensemble classifier. In this scenario, using EA improved the 3-model ensemble accuracy by 3.7%. However, when compared to the shared model with EA, the ensemble accuracy was 3.62% lower.
Evaluating the structure of cognitive tasks with transfer learning
Jul 28, 2023



Abstract:Electroencephalography (EEG) decoding is a challenging task due to the limited availability of labelled data. While transfer learning is a promising technique to address this challenge, it assumes that transferable data domains and task are known, which is not the case in this setting. This study investigates the transferability of deep learning representations between different EEG decoding tasks. We conduct extensive experiments using state-of-the-art decoding models on two recently released EEG datasets, ERP CORE and M$^3$CV, containing over 140 subjects and 11 distinct cognitive tasks. We measure the transferability of learned representations by pre-training deep neural networks on one task and assessing their ability to decode subsequent tasks. Our experiments demonstrate that, even with linear probing transfer, significant improvements in decoding performance can be obtained, with gains of up to 28% compare with the pure supervised approach. Additionally, we discover evidence that certain decoding paradigms elicit specific and narrow brain activities, while others benefit from pre-training on a broad range of representations. By revealing which tasks transfer well and demonstrating the benefits of transfer learning for EEG decoding, our findings have practical implications for mitigating data scarcity in this setting. The transfer maps generated also provide insights into the hierarchical relations between cognitive tasks, hence enhancing our understanding of how these tasks are connected from a neuroscientific standpoint.
End-to-end P300 BCI using Bayesian accumulation of Riemannian probabilities
Mar 15, 2022



Abstract:In brain-computer interfaces (BCI), most of the approaches based on event-related potential (ERP) focus on the detection of P300, aiming for single trial classification for a speller task. While this is an important objective, existing P300 BCI still require several repetitions to achieve a correct classification accuracy. Signal processing and machine learning advances in P300 BCI mostly revolve around the P300 detection part, leaving the character classification out of the scope. To reduce the number of repetitions while maintaining a good character classification, it is critical to embrace the full classification problem. We introduce an end-to-end pipeline, starting from feature extraction, and is composed of an ERP-level classification using probabilistic Riemannian MDM which feeds a character-level classification using Bayesian accumulation of confidence across trials. Whereas existing approaches only increase the confidence of a character when it is flashed, our new pipeline, called Bayesian accumulation of Riemannian probabilities (ASAP), update the confidence of each character after each flash. We provide the proper derivation and theoretical reformulation of this Bayesian approach for a seamless processing of information from signal to BCI characters. We demonstrate that our approach performs significantly better than standard methods on public P300 datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge