Raphael Y. de Camargo
Retrieval-Enhanced Named Entity Recognition
Oct 17, 2024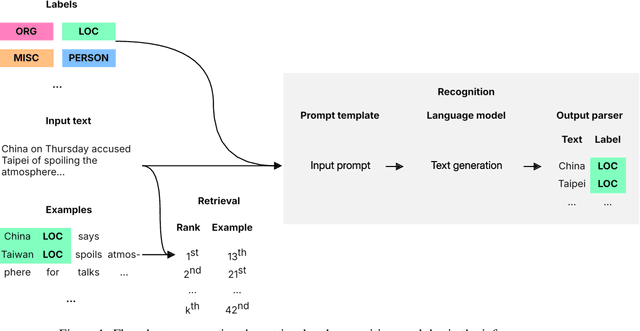
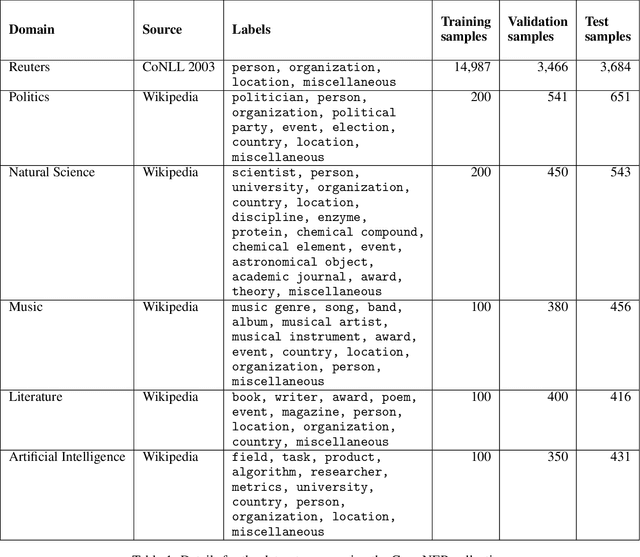

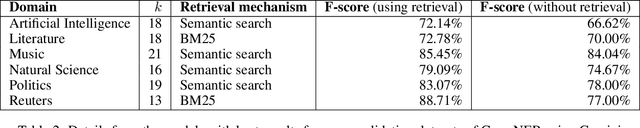
Abstract:When combined with In-Context Learning, a technique that enables models to adapt to new tasks by incorporating task-specific examples or demonstrations directly within the input prompt, autoregressive language models have achieved good performance in a wide range of tasks and applications. However, this combination has not been properly explored in the context of named entity recognition, where the structure of this task poses unique challenges. We propose RENER (Retrieval-Enhanced Named Entity Recognition), a technique for named entity recognition using autoregressive language models based on In-Context Learning and information retrieval techniques. When presented with an input text, RENER fetches similar examples from a dataset of training examples that are used to enhance a language model to recognize named entities from this input text. RENER is modular and independent of the underlying language model and information retrieval algorithms. Experimental results show that in the CrossNER collection we achieve state-of-the-art performance with the proposed technique and that information retrieval can increase the F-score by up to 11 percentage points.
Combining Euclidean Alignment and Data Augmentation for BCI decoding
May 23, 2024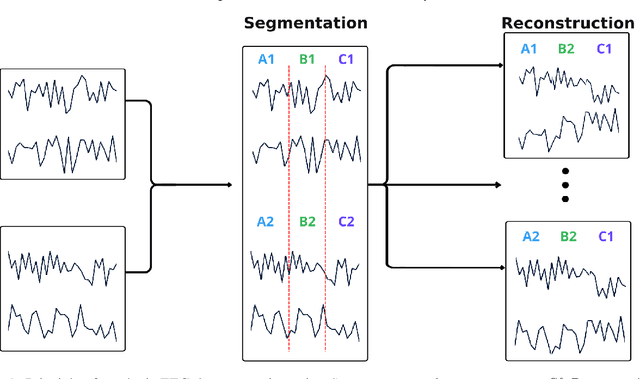
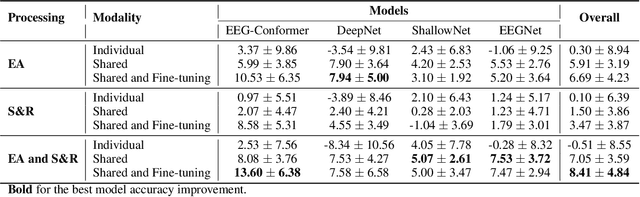
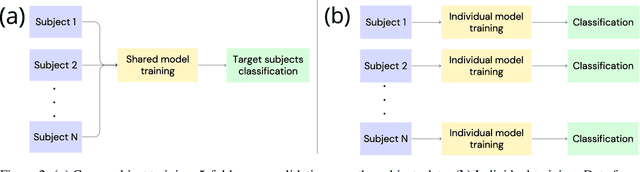
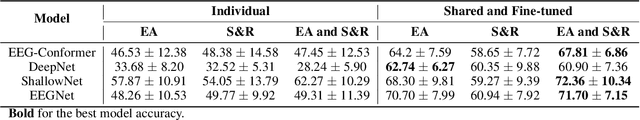
Abstract:Automated classification of electroencephalogram (EEG) signals is complex due to their high dimensionality, non-stationarity, low signal-to-noise ratio, and variability between subjects. Deep neural networks (DNNs) have shown promising results for EEG classification, but the above challenges hinder their performance. Euclidean Alignment (EA) and Data Augmentation (DA) are two promising techniques for improving DNN training by permitting the use of data from multiple subjects, increasing the data, and regularizing the available data. In this paper, we perform a detailed evaluation of the combined use of EA and DA with DNNs for EEG decoding. We trained individual models and shared models with data from multiple subjects and showed that combining EA and DA generates synergies that improve the accuracy of most models and datasets. Also, the shared models combined with fine-tuning benefited the most, with an overall increase of 8.41\% in classification accuracy.
Geometric Neural Network based on Phase Space for BCI decoding
Mar 08, 2024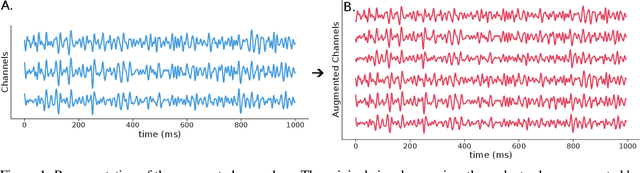
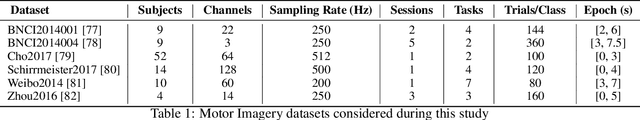
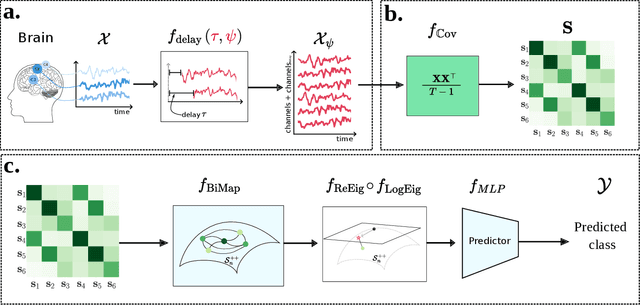
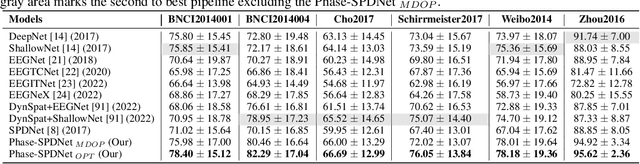
Abstract:The integration of Deep Learning (DL) algorithms on brain signal analysis is still in its nascent stages compared to their success in fields like Computer Vision, especially in Brain-Computer Interface (BCI), where the brain activity is decoded to control external devices without requiring muscle control. Electroencephalography (EEG) is a widely adopted choice for designing BCI systems due to its non-invasive and cost-effective nature and excellent temporal resolution. Still, it comes at the expense of limited training data, poor signal-to-noise, and a large variability across and within-subject recordings. Finally, setting up a BCI system with many electrodes takes a long time, hindering the widespread adoption of reliable DL architectures in BCIs outside research laboratories. To improve adoption, we need to improve user comfort using, for instance, reliable algorithms that operate with few electrodes. \textbf{Approach:} Our research aims to develop a DL algorithm that delivers effective results with a limited number of electrodes. Taking advantage of the Augmented Covariance Method with SPDNet, we propose the SPDNet$_{\psi}$ architecture and analyze its performance and computational impact, as well as the interpretability of the results. The evaluation is conducted on 5-fold cross-validation, using only three electrodes positioned above the Motor Cortex. The methodology was tested on nearly 100 subjects from several open-source datasets using the Mother Of All BCI Benchmark (MOABB) framework. \textbf{Main results:} The results of our SPDNet$_{\psi}$ demonstrate that the augmented approach combined with the SPDNet significantly outperforms all the current state-of-the-art DL architecture in MI decoding. \textbf{Significance:} This new architecture is explainable, with a low number of trainable parameters and a reduced carbon footprint.
A Systematic Evaluation of Euclidean Alignment with Deep Learning for EEG Decoding
Jan 30, 2024Abstract:Electroencephalography (EEG) signals are frequently used for various Brain-Computer Interface (BCI) tasks. While Deep Learning (DL) techniques have shown promising results, they are hindered by the substantial data requirements. By leveraging data from multiple subjects, transfer learning enables more effective training of DL models. A technique that is gaining popularity is Euclidean Alignment (EA) due to its ease of use, low computational complexity, and compatibility with Deep Learning models. However, few studies evaluate its impact on the training performance of shared and individual DL models. In this work, we systematically evaluate the effect of EA combined with DL for decoding BCI signals. We used EA to train shared models with data from multiple subjects and evaluated its transferability to new subjects. Our experimental results show that it improves decoding in the target subject by 4.33% and decreases convergence time by more than 70%. We also trained individual models for each subject to use as a majority-voting ensemble classifier. In this scenario, using EA improved the 3-model ensemble accuracy by 3.7%. However, when compared to the shared model with EA, the ensemble accuracy was 3.62% lower.
Evaluating the structure of cognitive tasks with transfer learning
Jul 28, 2023



Abstract:Electroencephalography (EEG) decoding is a challenging task due to the limited availability of labelled data. While transfer learning is a promising technique to address this challenge, it assumes that transferable data domains and task are known, which is not the case in this setting. This study investigates the transferability of deep learning representations between different EEG decoding tasks. We conduct extensive experiments using state-of-the-art decoding models on two recently released EEG datasets, ERP CORE and M$^3$CV, containing over 140 subjects and 11 distinct cognitive tasks. We measure the transferability of learned representations by pre-training deep neural networks on one task and assessing their ability to decode subsequent tasks. Our experiments demonstrate that, even with linear probing transfer, significant improvements in decoding performance can be obtained, with gains of up to 28% compare with the pure supervised approach. Additionally, we discover evidence that certain decoding paradigms elicit specific and narrow brain activities, while others benefit from pre-training on a broad range of representations. By revealing which tasks transfer well and demonstrating the benefits of transfer learning for EEG decoding, our findings have practical implications for mitigating data scarcity in this setting. The transfer maps generated also provide insights into the hierarchical relations between cognitive tasks, hence enhancing our understanding of how these tasks are connected from a neuroscientific standpoint.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge