Lukas Meyer
FruitNeRF++: A Generalized Multi-Fruit Counting Method Utilizing Contrastive Learning and Neural Radiance Fields
May 26, 2025



Abstract:We introduce FruitNeRF++, a novel fruit-counting approach that combines contrastive learning with neural radiance fields to count fruits from unstructured input photographs of orchards. Our work is based on FruitNeRF, which employs a neural semantic field combined with a fruit-specific clustering approach. The requirement for adaptation for each fruit type limits the applicability of the method, and makes it difficult to use in practice. To lift this limitation, we design a shape-agnostic multi-fruit counting framework, that complements the RGB and semantic data with instance masks predicted by a vision foundation model. The masks are used to encode the identity of each fruit as instance embeddings into a neural instance field. By volumetrically sampling the neural fields, we extract a point cloud embedded with the instance features, which can be clustered in a fruit-agnostic manner to obtain the fruit count. We evaluate our approach using a synthetic dataset containing apples, plums, lemons, pears, peaches, and mangoes, as well as a real-world benchmark apple dataset. Our results demonstrate that FruitNeRF++ is easier to control and compares favorably to other state-of-the-art methods.
Cherry Yield Forecast: Harvest Prediction for Individual Sweet Cherry Trees
Mar 26, 2025



Abstract:This paper is part of a publication series from the For5G project that has the goal of creating digital twins of sweet cherry trees. At the beginning a brief overview of the revious work in this project is provided. Afterwards the focus shifts to a crucial problem in the fruit farming domain: the difficulty of making reliable yield predictions early in the season. Following three Satin sweet cherry trees along the year 2023 enabled the collection of accurate ground truth data about the development of cherries from dormancy until harvest. The methodology used to collect this data is presented, along with its valuation and visualization. The predictive power of counting objects at all relevant vegetative stages of the fruit development cycle in cherry trees with regards to yield predictions is investigated. It is found that all investigated fruit states are suitable for yield predictions based on linear regression. Conceptionally, there is a trade-off between earliness and external events with the potential to invalidate the prediction. Considering this, two optimal timepoints are suggested that are opening cluster stage before the start of the flowering and the early fruit stage right after the second fruit drop. However, both timepoints are challenging to solve with automated procedures based on image data. Counting developing cherries based on images is exceptionally difficult due to the small fruit size and their tendency to be occluded by leaves. It was not possible to obtain satisfying results relying on a state-of-the-art fruit-counting method. Counting the elements within a bursting bud is also challenging, even when using high resolution cameras. It is concluded that accurate yield prediction for sweet cherry trees is possible when objects are manually counted and that automated features extraction with similar accuracy remains an open problem yet to be solved.
FruitNeRF: A Unified Neural Radiance Field based Fruit Counting Framework
Aug 12, 2024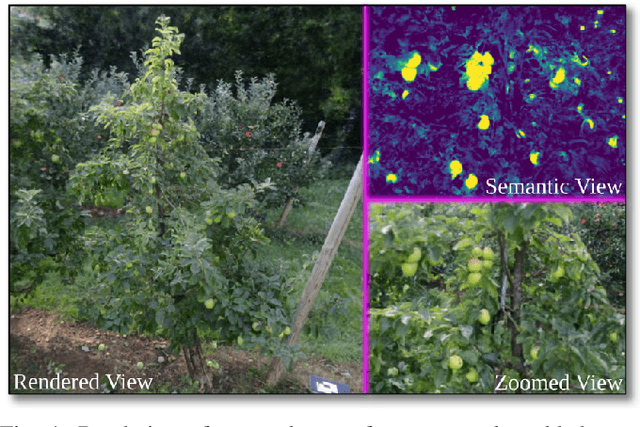
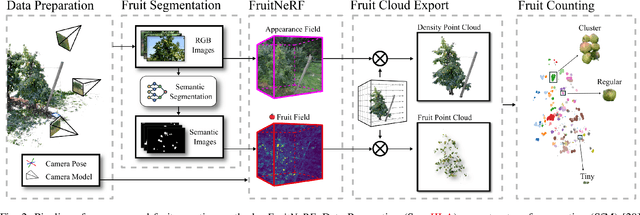
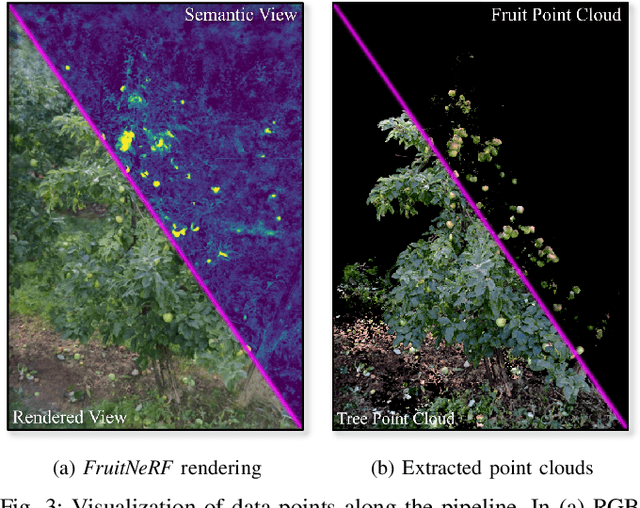
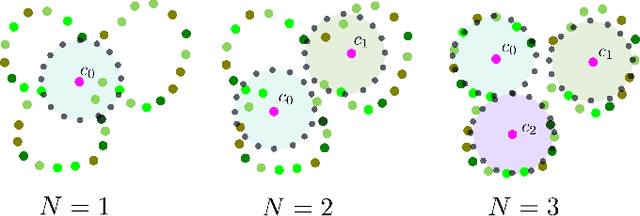
Abstract:We introduce FruitNeRF, a unified novel fruit counting framework that leverages state-of-the-art view synthesis methods to count any fruit type directly in 3D. Our framework takes an unordered set of posed images captured by a monocular camera and segments fruit in each image. To make our system independent of the fruit type, we employ a foundation model that generates binary segmentation masks for any fruit. Utilizing both modalities, RGB and semantic, we train a semantic neural radiance field. Through uniform volume sampling of the implicit Fruit Field, we obtain fruit-only point clouds. By applying cascaded clustering on the extracted point cloud, our approach achieves precise fruit count.The use of neural radiance fields provides significant advantages over conventional methods such as object tracking or optical flow, as the counting itself is lifted into 3D. Our method prevents double counting fruit and avoids counting irrelevant fruit.We evaluate our methodology using both real-world and synthetic datasets. The real-world dataset consists of three apple trees with manually counted ground truths, a benchmark apple dataset with one row and ground truth fruit location, while the synthetic dataset comprises various fruit types including apple, plum, lemon, pear, peach, and mango.Additionally, we assess the performance of fruit counting using the foundation model compared to a U-Net.
PEGASUS: Physically Enhanced Gaussian Splatting Simulation System for 6DOF Object Pose Dataset Generation
Jan 04, 2024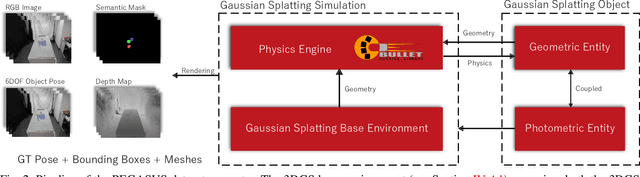


Abstract:We introduce Physically Enhanced Gaussian Splatting Simulation System (PEGASUS) for 6DOF object pose dataset generation, a versatile dataset generator based on 3D Gaussian Splatting. Environment and object representations can be easily obtained using commodity cameras to reconstruct with Gaussian Splatting. PEGASUS allows the composition of new scenes by merging the respective underlying Gaussian Splatting point cloud of an environment with one or multiple objects. Leveraging a physics engine enables the simulation of natural object placement within a scene through interaction between meshes extracted for the objects and the environment. Consequently, an extensive amount of new scenes - static or dynamic - can be created by combining different environments and objects. By rendering scenes from various perspectives, diverse data points such as RGB images, depth maps, semantic masks, and 6DoF object poses can be extracted. Our study demonstrates that training on data generated by PEGASUS enables pose estimation networks to successfully transfer from synthetic data to real-world data. Moreover, we introduce the Ramen dataset, comprising 30 Japanese cup noodle items. This dataset includes spherical scans that captures images from both object hemisphere and the Gaussian Splatting reconstruction, making them compatible with PEGASUS.
CherryPicker: Semantic Skeletonization and Topological Reconstruction of Cherry Trees
Apr 10, 2023



Abstract:In plant phenotyping, accurate trait extraction from 3D point clouds of trees is still an open problem. For automatic modeling and trait extraction of tree organs such as blossoms and fruits, the semantically segmented point cloud of a tree and the tree skeleton are necessary. Therefore, we present CherryPicker, an automatic pipeline that reconstructs photo-metric point clouds of trees, performs semantic segmentation and extracts their topological structure in form of a skeleton. Our system combines several state-of-the-art algorithms to enable automatic processing for further usage in 3D-plant phenotyping applications. Within this pipeline, we present a method to automatically estimate the scale factor of a monocular reconstruction to overcome scale ambiguity and obtain metrically correct point clouds. Furthermore, we propose a semantic skeletonization algorithm build up on Laplacian-based contraction. We also show by weighting different tree organs semantically, our approach can effectively remove artifacts induced by occlusion and structural size variations. CherryPicker obtains high-quality topology reconstructions of cherry trees with precise details.
ISLES 2022: A multi-center magnetic resonance imaging stroke lesion segmentation dataset
Jun 14, 2022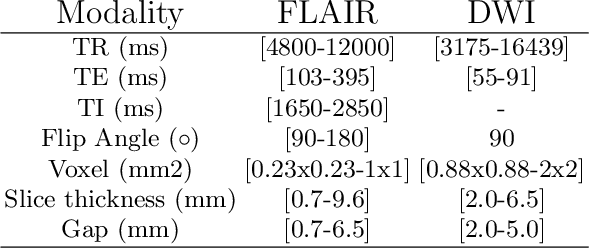
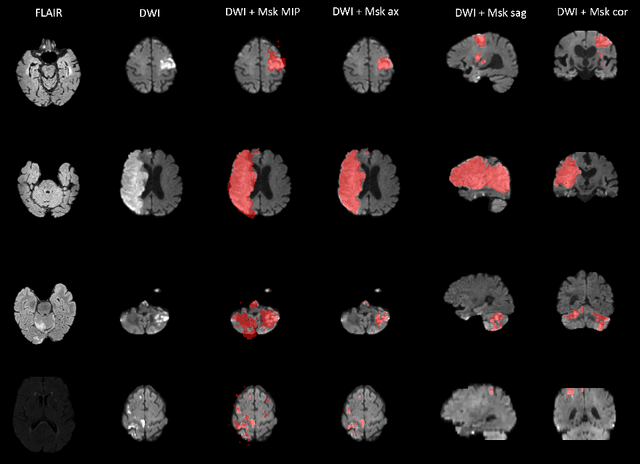

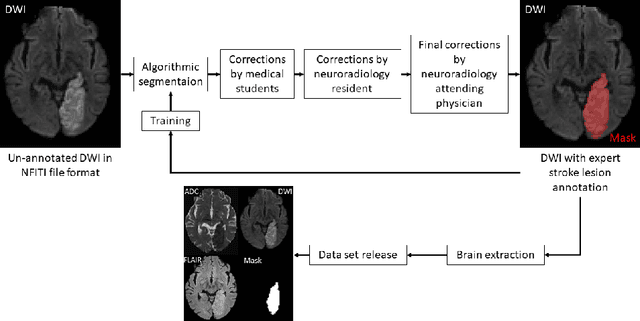
Abstract:Magnetic resonance imaging (MRI) is a central modality for stroke imaging. It is used upon patient admission to make treatment decisions such as selecting patients for intravenous thrombolysis or endovascular therapy. MRI is later used in the duration of hospital stay to predict outcome by visualizing infarct core size and location. Furthermore, it may be used to characterize stroke etiology, e.g. differentiation between (cardio)-embolic and non-embolic stroke. Computer based automated medical image processing is increasingly finding its way into clinical routine. Previous iterations of the Ischemic Stroke Lesion Segmentation (ISLES) challenge have aided in the generation of identifying benchmark methods for acute and sub-acute ischemic stroke lesion segmentation. Here we introduce an expert-annotated, multicenter MRI dataset for segmentation of acute to subacute stroke lesions. This dataset comprises 400 multi-vendor MRI cases with high variability in stroke lesion size, quantity and location. It is split into a training dataset of n=250 and a test dataset of n=150. All training data will be made publicly available. The test dataset will be used for model validation only and will not be released to the public. This dataset serves as the foundation of the ISLES 2022 challenge with the goal of finding algorithmic methods to enable the development and benchmarking of robust and accurate segmentation algorithms for ischemic stroke.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge