Lingze Zeng
NeurDB: An AI-powered Autonomous Data System
May 07, 2024Abstract:In the wake of rapid advancements in artificial intelligence (AI), we stand on the brink of a transformative leap in data systems. The imminent fusion of AI and DB (AIxDB) promises a new generation of data systems, which will relieve the burden on end-users across all industry sectors by featuring AI-enhanced functionalities, such as personalized and automated in-database AI-powered analytics, self-driving capabilities for improved system performance, etc. In this paper, we explore the evolution of data systems with a focus on deepening the fusion of AI and DB. We present NeurDB, our next-generation data system designed to fully embrace AI design in each major system component and provide in-database AI-powered analytics. We outline the conceptual and architectural overview of NeurDB, discuss its design choices and key components, and report its current development and future plan.
Powering In-Database Dynamic Model Slicing for Structured Data Analytics
May 01, 2024



Abstract:Relational database management systems (RDBMS) are widely used for the storage and retrieval of structured data. To derive insights beyond statistical aggregation, we typically have to extract specific subdatasets from the database using conventional database operations, and then apply deep neural networks (DNN) training and inference on these respective subdatasets in a separate machine learning system. The process can be prohibitively expensive, especially when there are a combinatorial number of subdatasets extracted for different analytical purposes. This calls for efficient in-database support of advanced analytical methods In this paper, we introduce LEADS, a novel SQL-aware dynamic model slicing technique to customize models for subdatasets specified by SQL queries. LEADS improves the predictive modeling of structured data via the mixture of experts (MoE) technique and maintains inference efficiency by a SQL-aware gating network. At the core of LEADS is the construction of a general model with multiple expert sub-models via MoE trained over the entire database. This SQL-aware MoE technique scales up the modeling capacity, enhances effectiveness, and preserves efficiency by activating only necessary experts via the gating network during inference. Additionally, we introduce two regularization terms during the training process of LEADS to strike a balance between effectiveness and efficiency. We also design and build an in-database inference system, called INDICES, to support end-to-end advanced structured data analytics by non-intrusively incorporating LEADS onto PostgreSQL. Our extensive experiments on real-world datasets demonstrate that LEADS consistently outperforms baseline models, and INDICES delivers effective in-database analytics with a considerable reduction in inference latency compared to traditional solutions.
Learning in Imperfect Environment: Multi-Label Classification with Long-Tailed Distribution and Partial Labels
Apr 20, 2023Abstract:Conventional multi-label classification (MLC) methods assume that all samples are fully labeled and identically distributed. Unfortunately, this assumption is unrealistic in large-scale MLC data that has long-tailed (LT) distribution and partial labels (PL). To address the problem, we introduce a novel task, Partial labeling and Long-Tailed Multi-Label Classification (PLT-MLC), to jointly consider the above two imperfect learning environments. Not surprisingly, we find that most LT-MLC and PL-MLC approaches fail to solve the PLT-MLC, resulting in significant performance degradation on the two proposed PLT-MLC benchmarks. Therefore, we propose an end-to-end learning framework: \textbf{CO}rrection $\rightarrow$ \textbf{M}odificat\textbf{I}on $\rightarrow$ balan\textbf{C}e, abbreviated as \textbf{\method{}}. Our bootstrapping philosophy is to simultaneously correct the missing labels (Correction) with convinced prediction confidence over a class-aware threshold and to learn from these recall labels during training. We next propose a novel multi-focal modifier loss that simultaneously addresses head-tail imbalance and positive-negative imbalance to adaptively modify the attention to different samples (Modification) under the LT class distribution. In addition, we develop a balanced training strategy by distilling the model's learning effect from head and tail samples, and thus design a balanced classifier (Balance) conditioned on the head and tail learning effect to maintain stable performance for all samples. Our experimental study shows that the proposed \method{} significantly outperforms general MLC, LT-MLC and PL-MLC methods in terms of effectiveness and robustness on our newly created PLT-MLC datasets.
Toward Cohort Intelligence: A Universal Cohort Representation Learning Framework for Electronic Health Record Analysis
Apr 12, 2023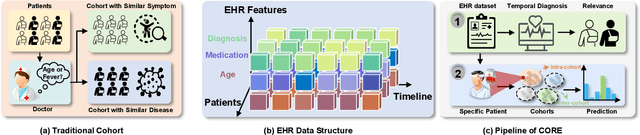
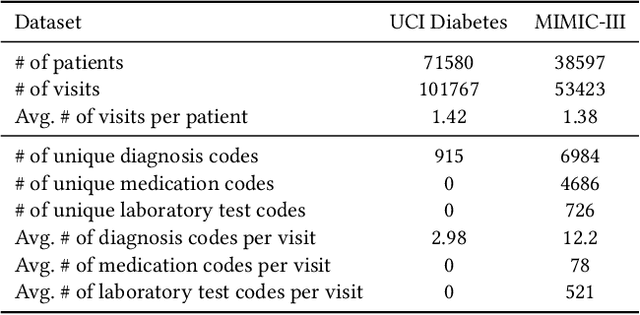
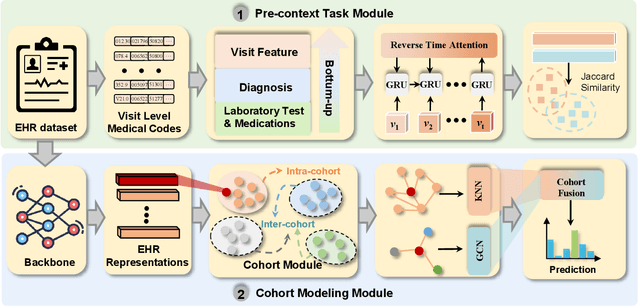
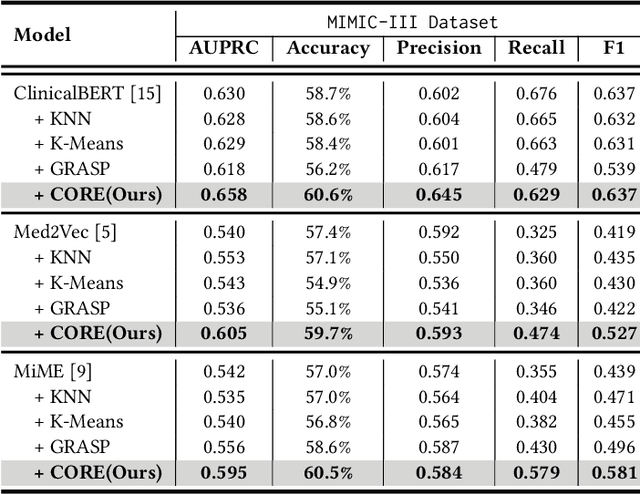
Abstract:Electronic Health Records (EHR) are generated from clinical routine care recording valuable information of broad patient populations, which provide plentiful opportunities for improving patient management and intervention strategies in clinical practice. To exploit the enormous potential of EHR data, a popular EHR data analysis paradigm in machine learning is EHR representation learning, which first leverages the individual patient's EHR data to learn informative representations by a backbone, and supports diverse health-care downstream tasks grounded on the representations. Unfortunately, such a paradigm fails to access the in-depth analysis of patients' relevance, which is generally known as cohort studies in clinical practice. Specifically, patients in the same cohort tend to share similar characteristics, implying their resemblance in medical conditions such as symptoms or diseases. In this paper, we propose a universal COhort Representation lEarning (CORE) framework to augment EHR utilization by leveraging the fine-grained cohort information among patients. In particular, CORE first develops an explicit patient modeling task based on the prior knowledge of patients' diagnosis codes, which measures the latent relevance among patients to adaptively divide the cohorts for each patient. Based on the constructed cohorts, CORE recodes the pre-extracted EHR data representation from intra- and inter-cohort perspectives, yielding augmented EHR data representation learning. CORE is readily applicable to diverse backbone models, serving as a universal plug-in framework to infuse cohort information into healthcare methods for boosted performance. We conduct an extensive experimental evaluation on two real-world datasets, and the experimental results demonstrate the effectiveness and generalizability of CORE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge