Krishna S. Nayak
A Feasibility Study of Task-Based fMRI at 0.55 T
May 26, 2025Abstract:0.55T MRI offers advantages compared to conventional field strengths, including reduced susceptibility artifacts and better compatibility with simultaneous EEG recordings. However, reliable task-based fMRI at 0.55T has not been significantly demonstrated. In this study, we establish a robust task-based fMRI protocol and analysis pipeline at 0.55T that achieves full brain coverage and results comparable to what is expected for activation extent and location. We attempted fMRI at 0.55T by combining EPI acquisition with custom analysis techniques. Finger-tapping and visual tasks were used, comparing 5- and 10-minute runs to enhance activation detection. The results show significant activations, demonstrating that high-quality task-based fMRI is achievable at 0.55T in single subjects. This study demonstrates that reliable task-based fMRI is feasible on 0.55T scanners, potentially broadening functional neuroimaging access in clinical and research settings where high-field MRI is unavailable or impractical, supporting broader diagnostic and research applications.
A multispeaker dataset of raw and reconstructed speech production real-time MRI video and 3D volumetric images
Feb 16, 2021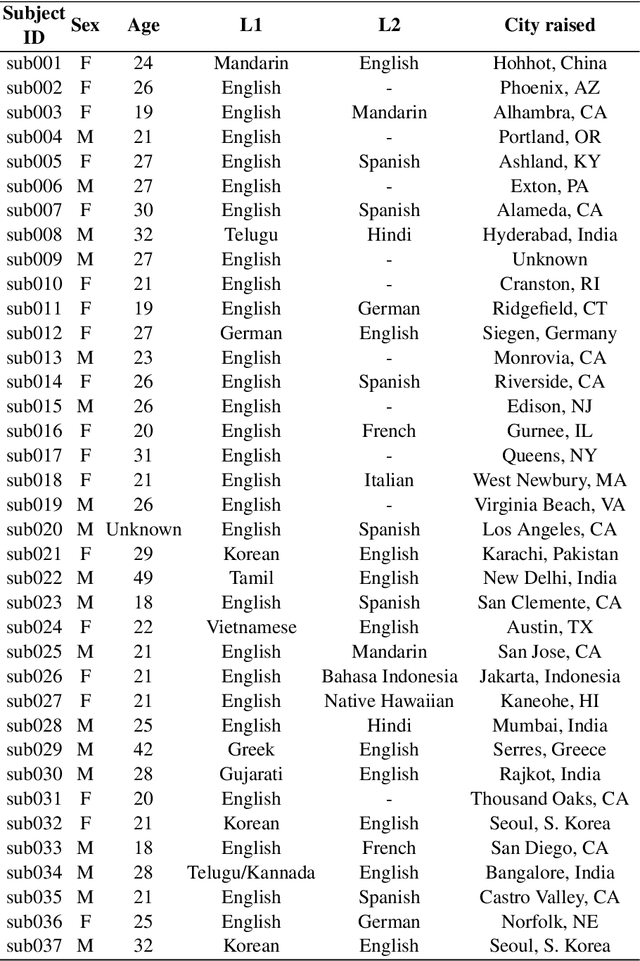
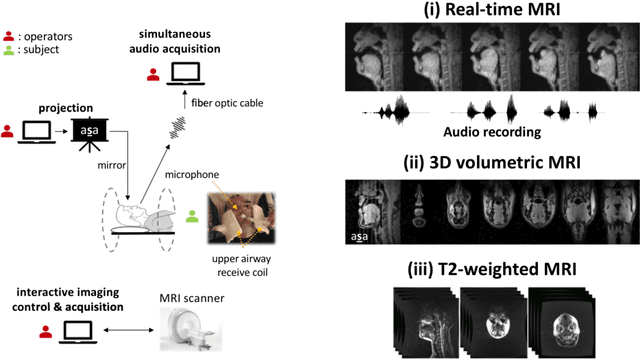
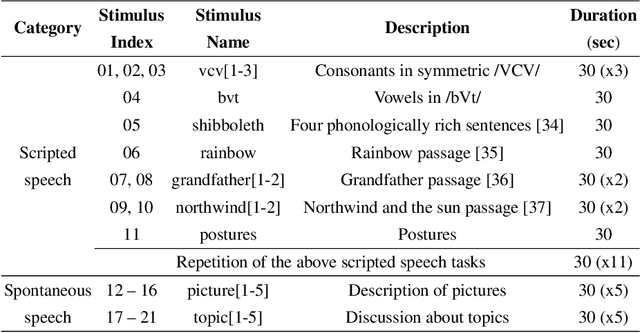
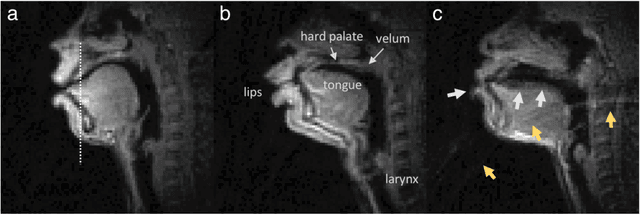
Abstract:Real-time magnetic resonance imaging (RT-MRI) of human speech production is enabling significant advances in speech science, linguistics, bio-inspired speech technology development, and clinical applications. Easy access to RT-MRI is however limited, and comprehensive datasets with broad access are needed to catalyze research across numerous domains. The imaging of the rapidly moving articulators and dynamic airway shaping during speech demands high spatio-temporal resolution and robust reconstruction methods. Further, while reconstructed images have been published, to-date there is no open dataset providing raw multi-coil RT-MRI data from an optimized speech production experimental setup. Such datasets could enable new and improved methods for dynamic image reconstruction, artifact correction, feature extraction, and direct extraction of linguistically-relevant biomarkers. The present dataset offers a unique corpus of 2D sagittal-view RT-MRI videos along with synchronized audio for 75 subjects performing linguistically motivated speech tasks, alongside the corresponding first-ever public domain raw RT-MRI data. The dataset also includes 3D volumetric vocal tract MRI during sustained speech sounds and high-resolution static anatomical T2-weighted upper airway MRI for each subject.
Attention-gated convolutional neural networks for off-resonance correction of spiral real-time MRI
Feb 14, 2021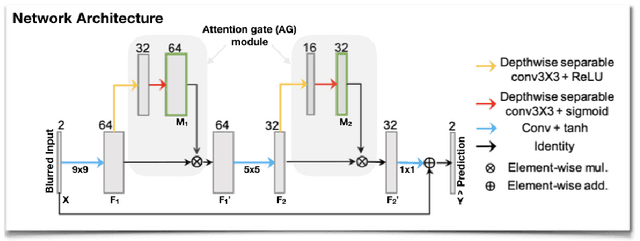
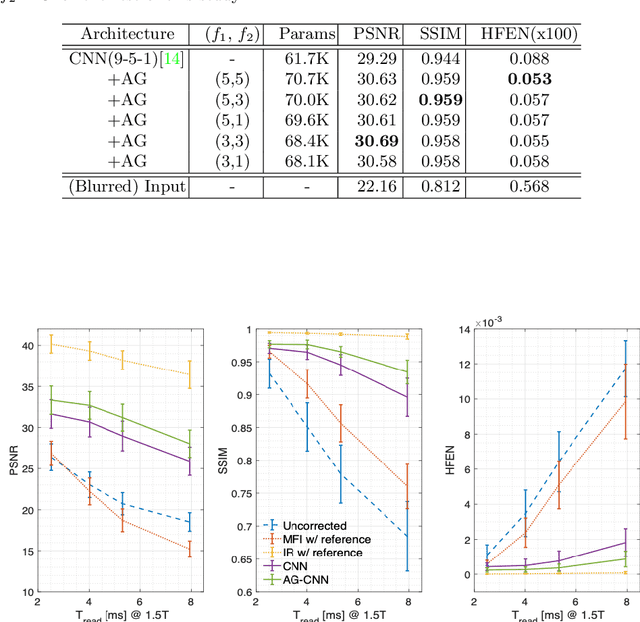

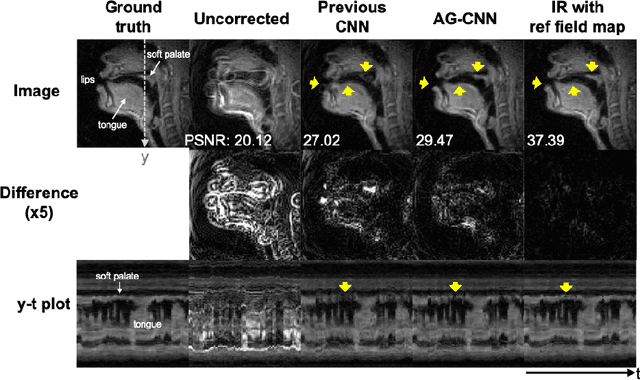
Abstract:Spiral acquisitions are preferred in real-time MRI because of their efficiency, which has made it possible to capture vocal tract dynamics during natural speech. A fundamental limitation of spirals is blurring and signal loss due to off-resonance, which degrades image quality at air-tissue boundaries. Here, we present a new CNN-based off-resonance correction method that incorporates an attention-gate mechanism. This leverages spatial and channel relationships of filtered outputs and improves the expressiveness of the networks. We demonstrate improved performance with the attention-gate, on 1.5 Tesla spiral speech RT-MRI, compared to existing off-resonance correction methods.
* 8 pages, 4 figures, 1 table
Robust Autocalibrated Structured Low-Rank EPI Ghost Correction
Jul 30, 2019Abstract:Purpose: We propose and evaluate a new structured low-rank method for EPI ghost correction called Robust Autocalibrated LORAKS (RAC-LORAKS). The method can be used to suppress EPI ghosts arising from the differences between different readout gradient polarities and/or the differences between different shots. It does not require conventional EPI navigator signals, and is robust to imperfect autocalibration data. Methods: Autocalibrated LORAKS is a previous structured low-rank method for EPI ghost correction that uses GRAPPA-type autocalibration data to enable high-quality ghost correction. This method works well when the autocalibration data is pristine, but performance degrades substantially when the autocalibration information is imperfect. RAC-LORAKS generalizes Autocalibrated LORAKS in two ways. First, it does not completely trust the information from autocalibration data, and instead considers the autocalibration and EPI data simultaneously when estimating low-rank matrix structure. And second, it uses complementary information from the autocalibration data to improve EPI reconstruction in a multi-contrast joint reconstruction framework. RAC-LORAKS is evaluated using simulations and in vivo data, and compared to state-of-the-art methods. Results: RAC-LORAKS is demonstrated to have good ghost elimination performance compared to state-of-the-art methods in several complicated acquisition scenarios (including gradient-echo brain imaging, diffusion-encoded brain imaging, and cardiac imaging). Conclusion: RAC-LORAKS provides effective suppression of EPI ghosts and is robust to imperfect autocalibration data.
Accuracy, Uncertainty, and Adaptability of Automatic Myocardial ASL Segmentation using Deep CNN
Dec 10, 2018
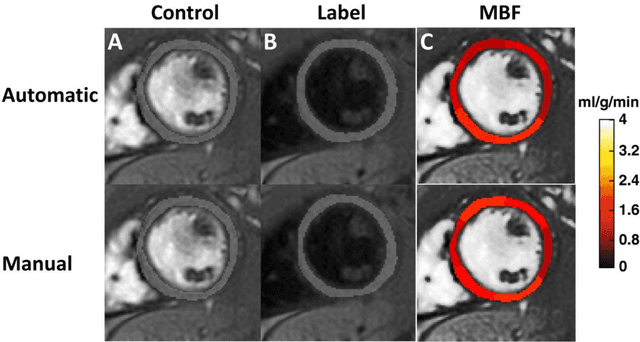


Abstract:PURPOSE: To apply deep convolutional neural networks (CNN) to the left ventricular segmentation task in myocardial arterial spin labeled (ASL) perfusion imaging. To develop methods that measure uncertainty and that adapt segmentation based on a specific false positive vs. false negative tradeoff. METHODS: We utilized a modified UNET architecture with Monte Carlo (MC) dropout. The model was trained on data from 22 subjects and tested on data from 6 heart transplant recipients. Manual segmentation and quantitative myocardial blood flow (MBF) maps were available for comparison. We consider two global scores of segmentation uncertainty, "Dice Uncertainty" and "MC Uncertainty", which were calculated with and without the use of manual segmentation, respectively. Tversky loss function with a hyperparameter $\beta$ was used to adapt the model to a specific false positive vs. false negative tradeoff. RESULTS: The modified UNET model achieved Dice coefficient of mean(std) = $0.91(0.04)$ on the test set. MBF measured using automatic segmentation was highly correlated to that measured using the manual segmentation ($R^2 = 0.96$). Dice Uncertainty and MC Uncertainty were in good agreement ($R^2 = 0.64$). As $\beta$ increased, the false positive rate systematically decreased and false negative rate systematically increased. CONCLUSION: We demonstrate the feasibility of using CNN for automatic segmentation of the left ventricle in myocardial ASL data. This is a particularly challenging application because of low and inconsistent blood-myocardium contrast-to-noise ratio. We also demonstrate novel methods that measure uncertainty and adapt to a desired tradeoff between false positive and false negative rates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge