Konstantin Voevodski
Monotonic Calibrated Interpolated Look-Up Tables
Jan 20, 2016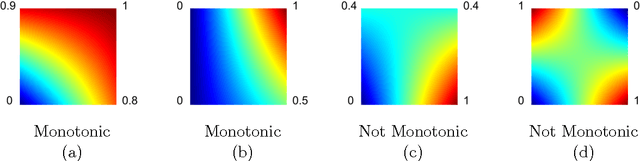
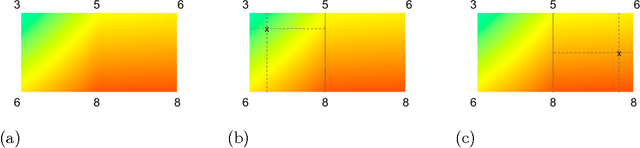
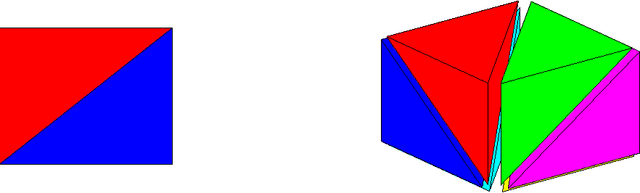
Abstract:Real-world machine learning applications may require functions that are fast-to-evaluate and interpretable. In particular, guaranteed monotonicity of the learned function can be critical to user trust. We propose meeting these goals for low-dimensional machine learning problems by learning flexible, monotonic functions using calibrated interpolated look-up tables. We extend the structural risk minimization framework of lattice regression to train monotonic look-up tables by solving a convex problem with appropriate linear inequality constraints. In addition, we propose jointly learning interpretable calibrations of each feature to normalize continuous features and handle categorical or missing data, at the cost of making the objective non-convex. We address large-scale learning through parallelization, mini-batching, and propose random sampling of additive regularizer terms. Case studies with real-world problems with five to sixteen features and thousands to millions of training samples demonstrate the proposed monotonic functions can achieve state-of-the-art accuracy on practical problems while providing greater transparency to users.
Local algorithms for interactive clustering
Mar 19, 2015



Abstract:We study the design of interactive clustering algorithms for data sets satisfying natural stability assumptions. Our algorithms start with any initial clustering and only make local changes in each step; both are desirable features in many applications. We show that in this constrained setting one can still design provably efficient algorithms that produce accurate clusterings. We also show that our algorithms perform well on real-world data.
Efficient Clustering with Limited Distance Information
Aug 09, 2014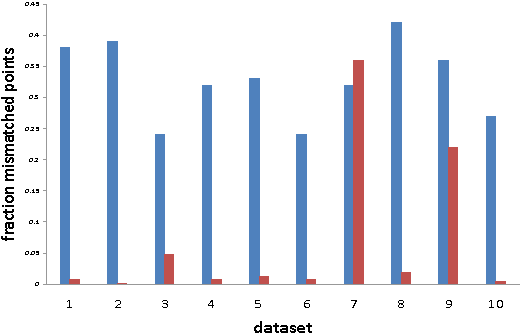
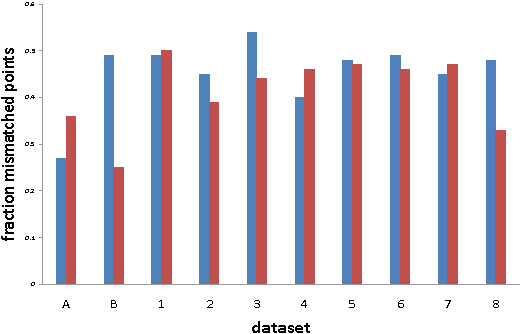
Abstract:Given a point set S and an unknown metric d on S, we study the problem of efficiently partitioning S into k clusters while querying few distances between the points. In our model we assume that we have access to one versus all queries that given a point s 2 S return the distances between s and all other points. We show that given a natural assumption about the structure of the instance, we can efficiently find an accurate clustering using only O(k) distance queries. We use our algorithm to cluster proteins by sequence similarity. This setting nicely fits our model because we can use a fast sequence database search program to query a sequence against an entire dataset. We conduct an empirical study that shows that even though we query a small fraction of the distances between the points, we produce clusterings that are close to a desired clustering given by manual classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge