Jun Ding
GCA-ResUNet: Medical Image Segmentation Using Grouped Coordinate Attention
Dec 30, 2025Abstract:Accurate segmentation of heterogeneous anatomical structures is pivotal for computer-aided diagnosis and subsequent clinical decision-making. Although U-Net based convolutional neural networks have achieved remarkable progress, their intrinsic locality and largely homogeneous attention formulations often limit the modeling of long-range contextual dependencies, especially in multi-organ scenarios and low-contrast regions. Transformer-based architectures mitigate this issue by leveraging global self-attention, but they usually require higher computational resources and larger training data, which may impede deployment in resource-constrained clinical environments.In this paper, we propose GCA-ResUNet, an efficient medical image segmentation framework equipped with a lightweight and plug-and-play Grouped Coordinate Attention (GCA) module. The proposed GCA decouples channel-wise context modeling into multiple groups to explicitly account for semantic heterogeneity across channels, and integrates direction-aware coordinate encoding to capture structured spatial dependencies along horizontal and vertical axes. This design enhances global representation capability while preserving the efficiency advantages of CNN backbones. Extensive experiments on two widely used benchmarks, Synapse and ACDC, demonstrate that GCA-ResUNet achieves Dice scores of 86.11% and 92.64%, respectively, outperforming a range of representative CNN and Transformer-based methods, including Swin-UNet and TransUNet. In particular, GCA-ResUNet yields consistent improvements in delineating small anatomical structures with complex boundaries. These results indicate that the proposed approach provides a favorable trade-off between segmentation accuracy and computational efficiency, offering a practical and scalable solution for clinical deployment.
GCA-ResUNet:Image segmentation in medical images using grouped coordinate attention
Nov 18, 2025Abstract:Medical image segmentation underpins computer-aided diagnosis and therapy by supporting clinical diagnosis, preoperative planning, and disease monitoring. While U-Net style convolutional neural networks perform well due to their encoder-decoder structures with skip connections, they struggle to capture long-range dependencies. Transformer-based variants address global context but often require heavy computation and large training datasets. This paper proposes GCA-ResUNet, an efficient segmentation network that integrates Grouped Coordinate Attention (GCA) into ResNet-50 residual blocks. GCA uses grouped coordinate modeling to jointly encode global dependencies across channels and spatial locations, strengthening feature representation and boundary delineation while adding minimal parameter and FLOP overhead compared with self-attention. On the Synapse dataset, GCA-ResUNet achieves a Dice score of 86.11%, and on the ACDC dataset, it reaches 92.64%, surpassing several state-of-the-art baselines while maintaining fast inference and favorable computational efficiency. These results indicate that GCA offers a practical way to enhance convolutional architectures with global modeling capability, enabling high-accuracy and resource-efficient medical image segmentation.
SUICA: Learning Super-high Dimensional Sparse Implicit Neural Representations for Spatial Transcriptomics
Dec 02, 2024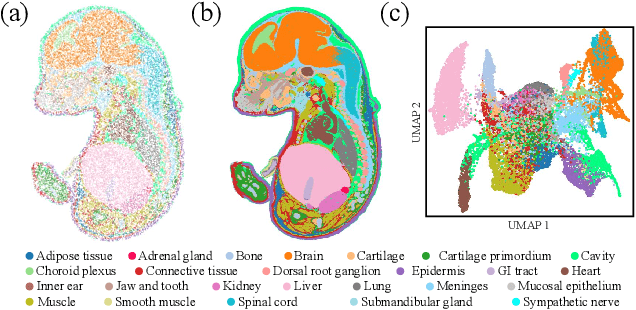
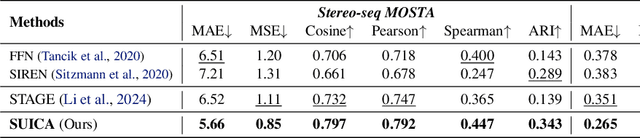
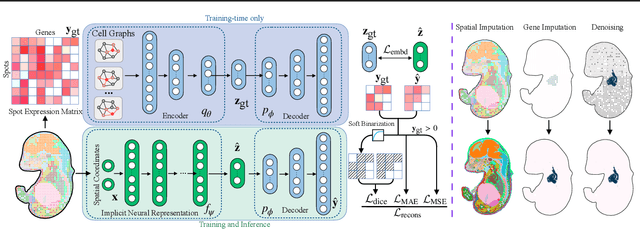
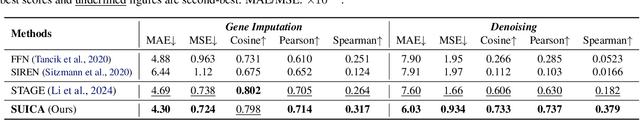
Abstract:Spatial Transcriptomics (ST) is a method that captures spatial gene expression profiles within histological sections. The discrete spatial distribution and the super-high dimensional sequencing results make ST data challenging to be modeled effectively. In this paper, we manage to model ST in a continuous and compact manner by the proposed tool, SUICA, empowered by the great approximation capability of Implicit Neural Representations (INRs) that can improve both the spatial resolution and the gene expression. Concretely within the proposed SUICA, we incorporate a graph-augmented Autoencoder to effectively model the context information of the unstructured spots and provide informative embeddings that are structure-aware for spatial mapping. We also tackle the extremely skewed distribution in a regression-by-classification fashion and enforce classification-based loss functions for the optimization of SUICA. By extensive experiments of a wide range of common ST platforms, SUICA outperforms both conventional INR variants and SOTA methods for ST super-resolution regarding numerical fidelity, statistical correlation, and bio-conservation. The prediction by SUICA also showcases amplified gene signatures that enriches the bio-conservation of the raw data and benefits subsequent analysis. The code is available at https://github.com/Szym29/SUICA.
Predictive Modeling of Flexible EHD Pumps using Kolmogorov-Arnold Networks
May 13, 2024


Abstract:We present a novel approach to predicting the pressure and flow rate of flexible electrohydrodynamic pumps using the Kolmogorov-Arnold Network. Inspired by the Kolmogorov-Arnold representation theorem, KAN replaces fixed activation functions with learnable spline-based activation functions, enabling it to approximate complex nonlinear functions more effectively than traditional models like Multi-Layer Perceptron and Random Forest. We evaluated KAN on a dataset of flexible EHD pump parameters and compared its performance against RF, and MLP models. KAN achieved superior predictive accuracy, with Mean Squared Errors of 12.186 and 0.001 for pressure and flow rate predictions, respectively. The symbolic formulas extracted from KAN provided insights into the nonlinear relationships between input parameters and pump performance. These findings demonstrate that KAN offers exceptional accuracy and interpretability, making it a promising alternative for predictive modeling in electrohydrodynamic pumping.
scBeacon: single-cell biomarker extraction via identifying paired cell clusters across biological conditions with contrastive siamese networks
Nov 05, 2023



Abstract:Despite the breakthroughs in biomarker discovery facilitated by differential gene analysis, challenges remain, particularly at the single-cell level. Traditional methodologies heavily rely on user-supplied cell annotations, focusing on individually expressed data, often neglecting the critical interactions between biological conditions, such as healthy versus diseased states. In response, here we introduce scBeacon, an innovative framework built upon a deep contrastive siamese network. scBeacon pioneers an unsupervised approach, adeptly identifying matched cell populations across varied conditions, enabling a refined differential gene analysis. By utilizing a VQ-VAE framework, a contrastive siamese network, and a greedy iterative strategy, scBeacon effectively pinpoints differential genes that hold potential as key biomarkers. Comprehensive evaluations on a diverse array of datasets validate scBeacon's superiority over existing single-cell differential gene analysis tools. Its precision and adaptability underscore its significant role in enhancing diagnostic accuracy in biomarker discovery. With the emphasis on the importance of biomarkers in diagnosis, scBeacon is positioned to be a pivotal asset in the evolution of personalized medicine and targeted treatments.
A Freeform Dielectric Metasurface Modeling Approach Based on Deep Neural Networks
Jan 01, 2020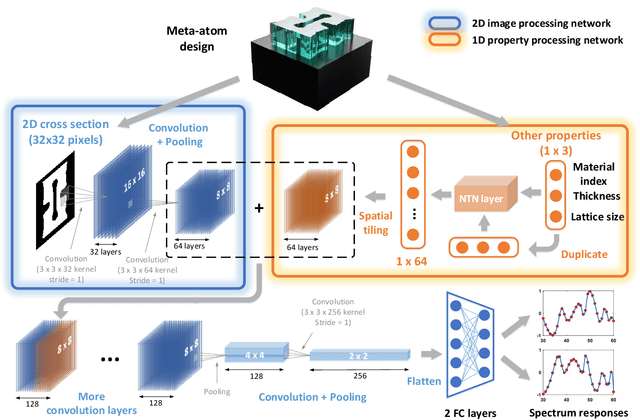
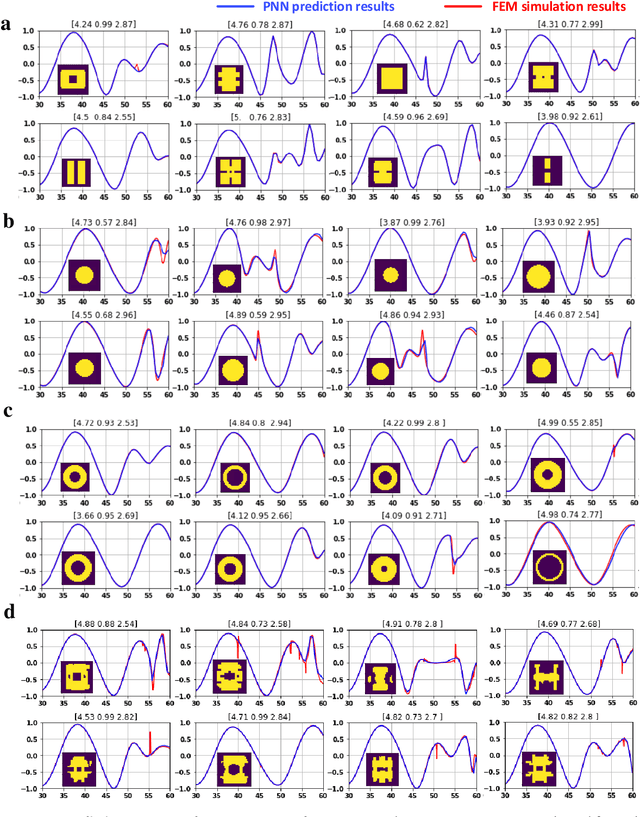
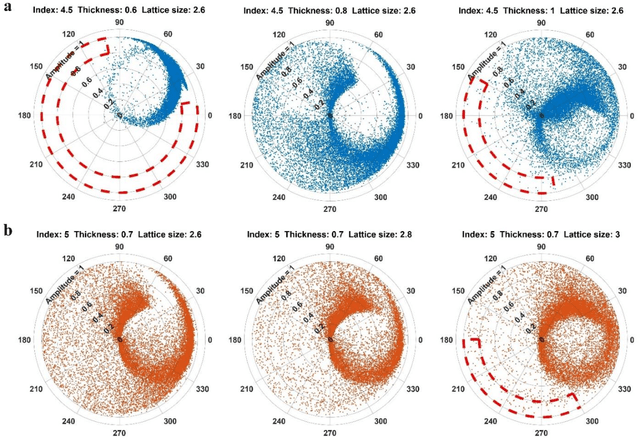
Abstract:Metasurfaces have shown promising potentials in shaping optical wavefronts while remaining compact compared to bulky geometric optics devices. Design of meta-atoms, the fundamental building blocks of metasurfaces, relies on trial-and-error method to achieve target electromagnetic responses. This process includes the characterization of an enormous amount of different meta-atom designs with different physical and geometric parameters, which normally demands huge computational resources. In this paper, a deep learning-based metasurface/meta-atom modeling approach is introduced to significantly reduce the characterization time while maintaining accuracy. Based on a convolutional neural network (CNN) structure, the proposed deep learning network is able to model meta-atoms with free-form 2D patterns and different lattice sizes, material refractive indexes and thicknesses. Moreover, the presented approach features the capability to predict meta-atoms' wide spectrum responses in the timescale of milliseconds, which makes it attractive for applications such as fast meta-atom/metasurface on-demand designs and optimizations.
A Novel Modeling Approach for All-Dielectric Metasurfaces Using Deep Neural Networks
Jun 08, 2019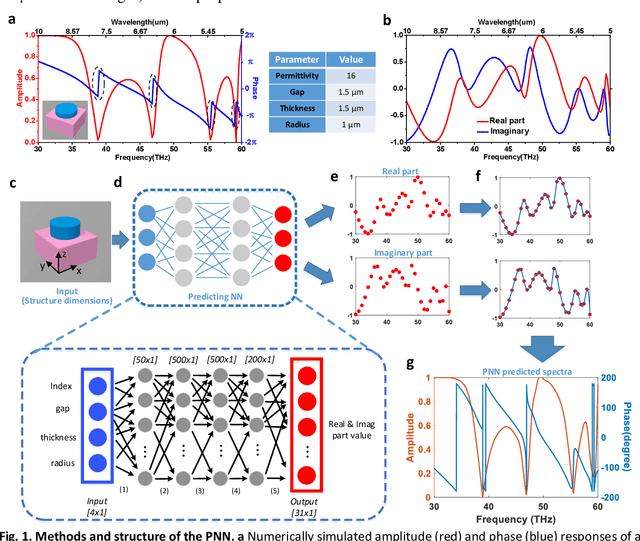
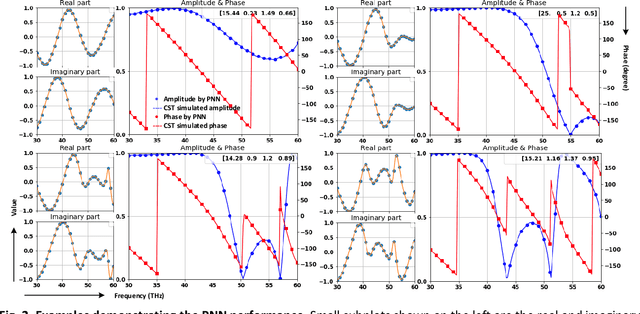
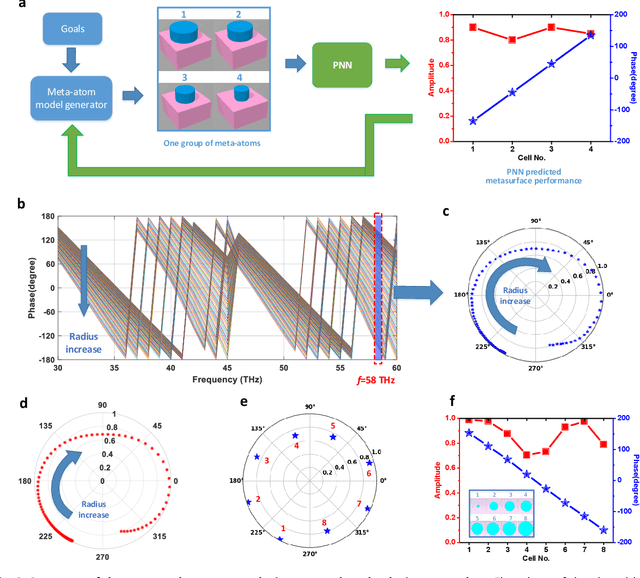
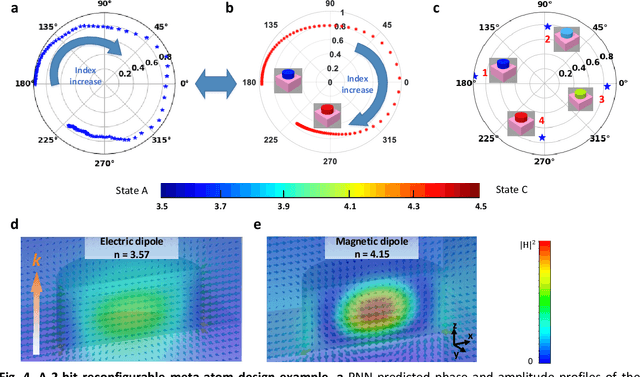
Abstract:Metasurfaces have become a promising means for manipulating optical wavefronts in flat and high-performance optical devices. Conventional metasurface device design relies on trial-and-error methods to obtain target electromagnetic (EM) response, an approach that demands significant efforts to investigate the enormous number of possible meta-atom structures. In this paper, a deep neural network approach is introduced that significantly improves on both speed and accuracy compared to techniques currently used to assemble metasurface-based devices. Our neural network approach overcomes three key challenges that have limited previous neural-network-based design schemes: input/output vector dimensional mismatch, accurate EM-wave phase prediction, as well as adaptation to 3-D dielectric structures, and can be generically applied to a wide variety of metasurface device designs across the entire electromagnetic spectrum. Using this new methodology, examples of neural networks capable of producing on-demand designs for meta-atoms, metasurface filters, and phase-change reconfigurable metasurfaces are demonstrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge