John R Hurst
A data-centric deep learning approach to airway segmentation
Jul 29, 2023Abstract:The morphology and distribution of airway tree abnormalities enables diagnosis and disease characterisation across a variety of chronic respiratory conditions. In this regard, airway segmentation plays a critical role in the production of the outline of the entire airway tree to enable estimation of disease extent and severity. In this study, we propose a data-centric deep learning technique to segment the airway tree. The proposed technique utilises interpolation and image split to improve data usefulness and quality. Then, an ensemble learning strategy is implemented to aggregate the segmented airway trees at different scales. In terms of segmentation performance (dice similarity coefficient), our method outperforms the baseline model by 2.5% on average when a combined loss is used. Further, our proposed technique has a low GPU usage and high flexibility enabling it to be deployed on any 2D deep learning model.
Airway measurement by refinement of synthetic images improves mortality prediction in idiopathic pulmonary fibrosis
Aug 30, 2022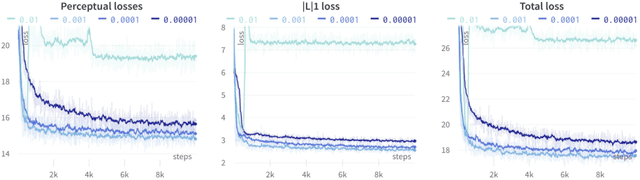
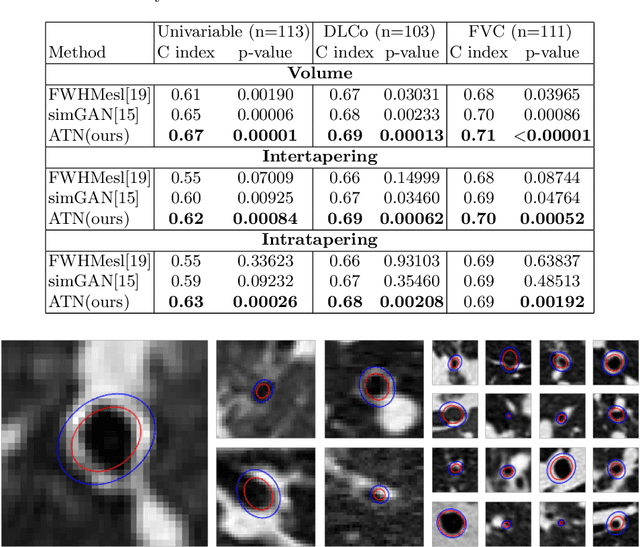
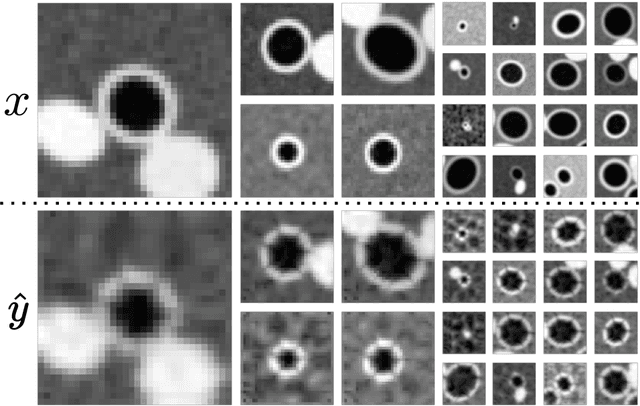
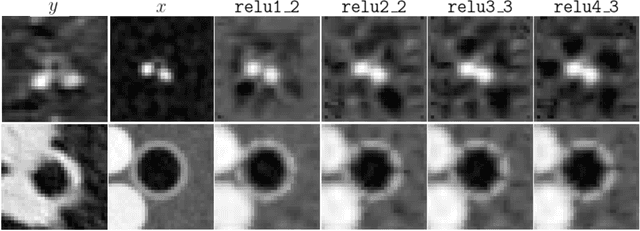
Abstract:Several chronic lung diseases, like idiopathic pulmonary fibrosis (IPF) are characterised by abnormal dilatation of the airways. Quantification of airway features on computed tomography (CT) can help characterise disease progression. Physics based airway measurement algorithms have been developed, but have met with limited success in part due to the sheer diversity of airway morphology seen in clinical practice. Supervised learning methods are also not feasible due to the high cost of obtaining precise airway annotations. We propose synthesising airways by style transfer using perceptual losses to train our model, Airway Transfer Network (ATN). We compare our ATN model with a state-of-the-art GAN-based network (simGAN) using a) qualitative assessment; b) assessment of the ability of ATN and simGAN based CT airway metrics to predict mortality in a population of 113 patients with IPF. ATN was shown to be quicker and easier to train than simGAN. ATN-based airway measurements were also found to be consistently stronger predictors of mortality than simGAN-derived airway metrics on IPF CTs. Airway synthesis by a transformation network that refines synthetic data using perceptual losses is a realistic alternative to GAN-based methods for clinical CT analyses of idiopathic pulmonary fibrosis. Our source code can be found at https://github.com/ashkanpakzad/ATN that is compatible with the existing open-source airway analysis framework, AirQuant.
Evaluation of automated airway morphological quantification for assessing fibrosing lung disease
Nov 19, 2021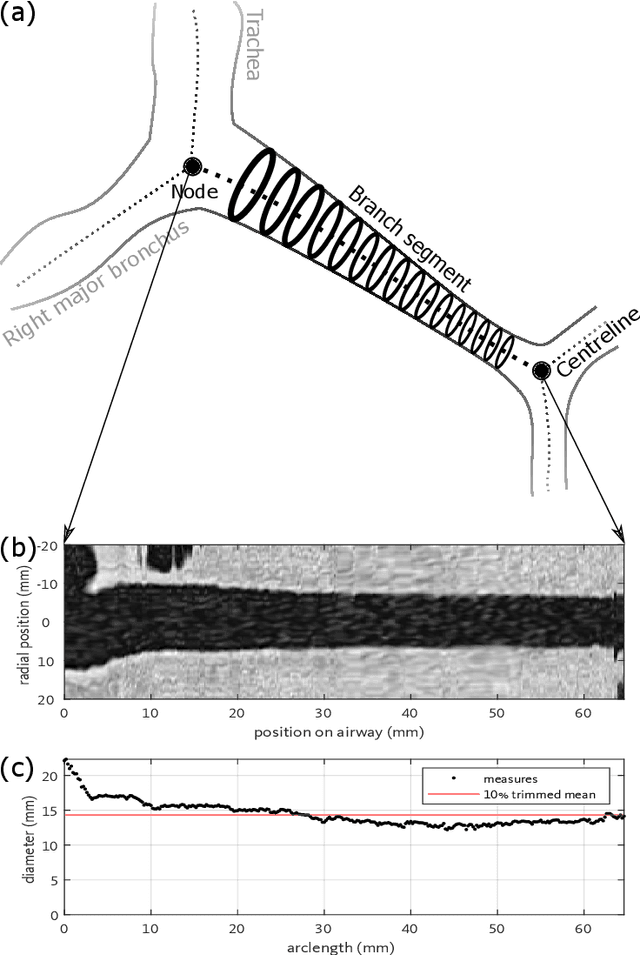
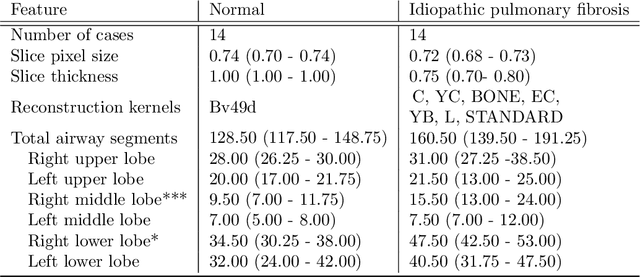
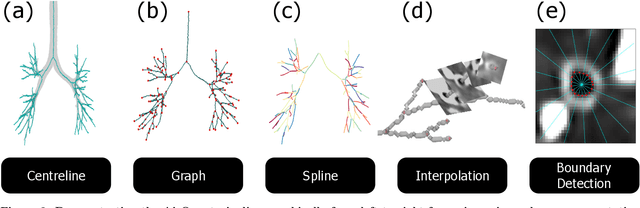
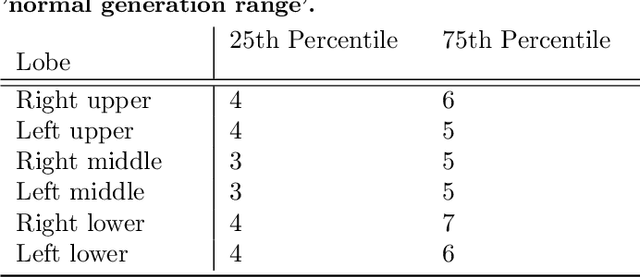
Abstract:Abnormal airway dilatation, termed traction bronchiectasis, is a typical feature of idiopathic pulmonary fibrosis (IPF). Volumetric computed tomography (CT) imaging captures the loss of normal airway tapering in IPF. We postulated that automated quantification of airway abnormalities could provide estimates of IPF disease extent and severity. We propose AirQuant, an automated computational pipeline that systematically parcellates the airway tree into its lobes and generational branches from a deep learning based airway segmentation, deriving airway structural measures from chest CT. Importantly, AirQuant prevents the occurrence of spurious airway branches by thick wave propagation and removes loops in the airway-tree by graph search, overcoming limitations of existing airway skeletonisation algorithms. Tapering between airway segments (intertapering) and airway tortuosity computed by AirQuant were compared between 14 healthy participants and 14 IPF patients. Airway intertapering was significantly reduced in IPF patients, and airway tortuosity was significantly increased when compared to healthy controls. Differences were most marked in the lower lobes, conforming to the typical distribution of IPF-related damage. AirQuant is an open-source pipeline that avoids limitations of existing airway quantification algorithms and has clinical interpretability. Automated airway measurements may have potential as novel imaging biomarkers of IPF severity and disease extent.
The challenges of deploying artificial intelligence models in a rapidly evolving pandemic
May 19, 2020Abstract:The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2, emerged into a world being rapidly transformed by artificial intelligence (AI) based on big data, computational power and neural networks. The gaze of these networks has in recent years turned increasingly towards applications in healthcare. It was perhaps inevitable that COVID-19, a global disease propagating health and economic devastation, should capture the attention and resources of the world's computer scientists in academia and industry. The potential for AI to support the response to the pandemic has been proposed across a wide range of clinical and societal challenges, including disease forecasting, surveillance and antiviral drug discovery. This is likely to continue as the impact of the pandemic unfolds on the world's people, industries and economy but a surprising observation on the current pandemic has been the limited impact AI has had to date in the management of COVID-19. This correspondence focuses on exploring potential reasons behind the lack of successful adoption of AI models developed for COVID-19 diagnosis and prognosis, in front-line healthcare services. We highlight the moving clinical needs that models have had to address at different stages of the epidemic, and explain the importance of translating models to reflect local healthcare environments. We argue that both basic and applied research are essential to accelerate the potential of AI models, and this is particularly so during a rapidly evolving pandemic. This perspective on the response to COVID-19, may provide a glimpse into how the global scientific community should react to combat future disease outbreaks more effectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge