Jing Xia
FedDEAP: Adaptive Dual-Prompt Tuning for Multi-Domain Federated Learning
Oct 21, 2025Abstract:Federated learning (FL) enables multiple clients to collaboratively train machine learning models without exposing local data, balancing performance and privacy. However, domain shift and label heterogeneity across clients often hinder the generalization of the aggregated global model. Recently, large-scale vision-language models like CLIP have shown strong zero-shot classification capabilities, raising the question of how to effectively fine-tune CLIP across domains in a federated setting. In this work, we propose an adaptive federated prompt tuning framework, FedDEAP, to enhance CLIP's generalization in multi-domain scenarios. Our method includes the following three key components: (1) To mitigate the loss of domain-specific information caused by label-supervised tuning, we disentangle semantic and domain-specific features in images by using semantic and domain transformation networks with unbiased mappings; (2) To preserve domain-specific knowledge during global prompt aggregation, we introduce a dual-prompt design with a global semantic prompt and a local domain prompt to balance shared and personalized information; (3) To maximize the inclusion of semantic and domain information from images in the generated text features, we align textual and visual representations under the two learned transformations to preserve semantic and domain consistency. Theoretical analysis and extensive experiments on four datasets demonstrate the effectiveness of our method in enhancing the generalization of CLIP for federated image recognition across multiple domains.
Serving Large Language Models on Huawei CloudMatrix384
Jun 15, 2025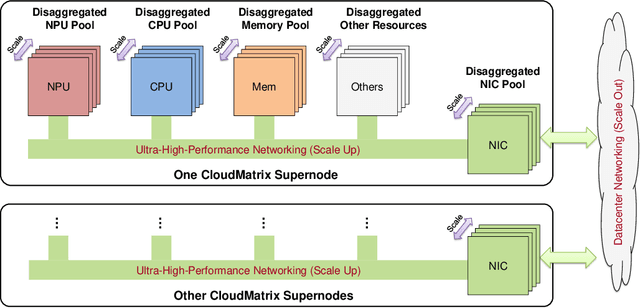
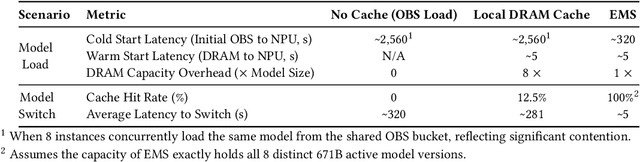
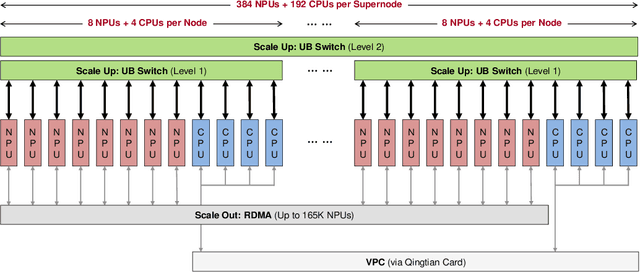
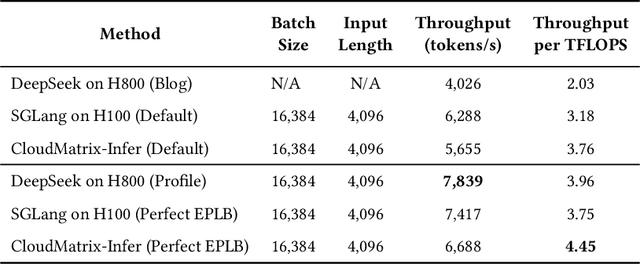
Abstract:The rapid evolution of large language models (LLMs), driven by growing parameter scales, adoption of mixture-of-experts (MoE) architectures, and expanding context lengths, imposes unprecedented demands on AI infrastructure. Traditional AI clusters face limitations in compute intensity, memory bandwidth, inter-chip communication, and latency, compounded by variable workloads and strict service-level objectives. Addressing these issues requires fundamentally redesigned hardware-software integration. This paper introduces Huawei CloudMatrix, a next-generation AI datacenter architecture, realized in the production-grade CloudMatrix384 supernode. It integrates 384 Ascend 910C NPUs and 192 Kunpeng CPUs interconnected via an ultra-high-bandwidth Unified Bus (UB) network, enabling direct all-to-all communication and dynamic pooling of resources. These features optimize performance for communication-intensive operations, such as large-scale MoE expert parallelism and distributed key-value cache access. To fully leverage CloudMatrix384, we propose CloudMatrix-Infer, an advanced LLM serving solution incorporating three core innovations: a peer-to-peer serving architecture that independently scales prefill, decode, and caching; a large-scale expert parallelism strategy supporting EP320 via efficient UB-based token dispatch; and hardware-aware optimizations including specialized operators, microbatch-based pipelining, and INT8 quantization. Evaluation with the DeepSeek-R1 model shows CloudMatrix-Infer achieves state-of-the-art efficiency: prefill throughput of 6,688 tokens/s per NPU and decode throughput of 1,943 tokens/s per NPU (<50 ms TPOT). It effectively balances throughput and latency, sustaining 538 tokens/s even under stringent 15 ms latency constraints, while INT8 quantization maintains model accuracy across benchmarks.
Bridging the Inter-Domain Gap through Low-Level Features for Cross-Modal Medical Image Segmentation
May 17, 2025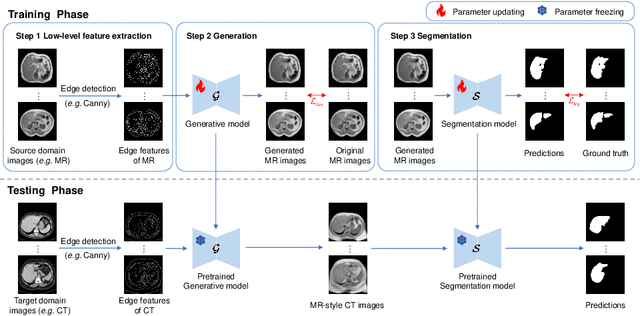
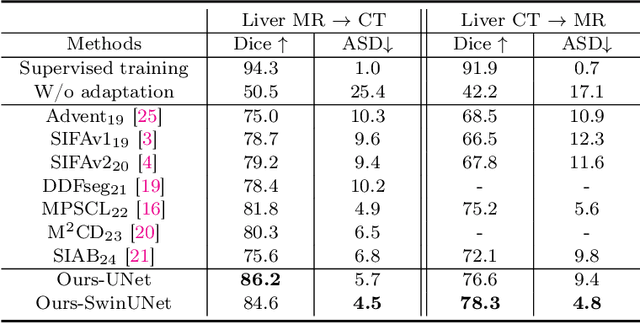
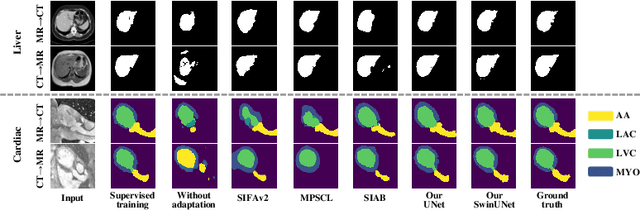
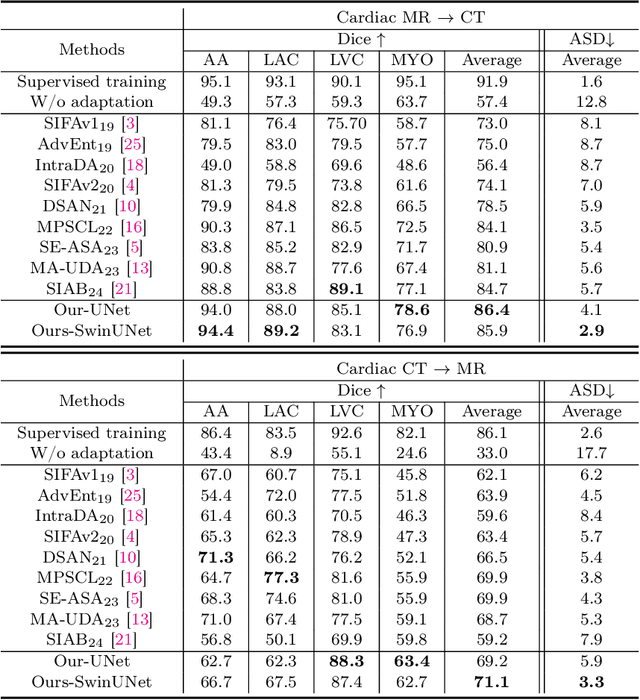
Abstract:This paper addresses the task of cross-modal medical image segmentation by exploring unsupervised domain adaptation (UDA) approaches. We propose a model-agnostic UDA framework, LowBridge, which builds on a simple observation that cross-modal images share some similar low-level features (e.g., edges) as they are depicting the same structures. Specifically, we first train a generative model to recover the source images from their edge features, followed by training a segmentation model on the generated source images, separately. At test time, edge features from the target images are input to the pretrained generative model to generate source-style target domain images, which are then segmented using the pretrained segmentation network. Despite its simplicity, extensive experiments on various publicly available datasets demonstrate that \proposed achieves state-of-the-art performance, outperforming eleven existing UDA approaches under different settings. Notably, further ablation studies show that \proposed is agnostic to different types of generative and segmentation models, suggesting its potential to be seamlessly plugged with the most advanced models to achieve even more outstanding results in the future. The code is available at https://github.com/JoshuaLPF/LowBridge.
Discovering robust biomarkers of neurological disorders from functional MRI using graph neural networks: A Review
May 01, 2024



Abstract:Graph neural networks (GNN) have emerged as a popular tool for modelling functional magnetic resonance imaging (fMRI) datasets. Many recent studies have reported significant improvements in disorder classification performance via more sophisticated GNN designs and highlighted salient features that could be potential biomarkers of the disorder. In this review, we provide an overview of how GNN and model explainability techniques have been applied on fMRI datasets for disorder prediction tasks, with a particular emphasis on the robustness of biomarkers produced for neurodegenerative diseases and neuropsychiatric disorders. We found that while most studies have performant models, salient features highlighted in these studies vary greatly across studies on the same disorder and little has been done to evaluate their robustness. To address these issues, we suggest establishing new standards that are based on objective evaluation metrics to determine the robustness of these potential biomarkers. We further highlight gaps in the existing literature and put together a prediction-attribution-evaluation framework that could set the foundations for future research on improving the robustness of potential biomarkers discovered via GNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge