Jiacheng Miao
Paper2Agent: Reimagining Research Papers As Interactive and Reliable AI Agents
Sep 08, 2025Abstract:We introduce Paper2Agent, an automated framework that converts research papers into AI agents. Paper2Agent transforms research output from passive artifacts into active systems that can accelerate downstream use, adoption, and discovery. Conventional research papers require readers to invest substantial effort to understand and adapt a paper's code, data, and methods to their own work, creating barriers to dissemination and reuse. Paper2Agent addresses this challenge by automatically converting a paper into an AI agent that acts as a knowledgeable research assistant. It systematically analyzes the paper and the associated codebase using multiple agents to construct a Model Context Protocol (MCP) server, then iteratively generates and runs tests to refine and robustify the resulting MCP. These paper MCPs can then be flexibly connected to a chat agent (e.g. Claude Code) to carry out complex scientific queries through natural language while invoking tools and workflows from the original paper. We demonstrate Paper2Agent's effectiveness in creating reliable and capable paper agents through in-depth case studies. Paper2Agent created an agent that leverages AlphaGenome to interpret genomic variants and agents based on ScanPy and TISSUE to carry out single-cell and spatial transcriptomics analyses. We validate that these paper agents can reproduce the original paper's results and can correctly carry out novel user queries. By turning static papers into dynamic, interactive AI agents, Paper2Agent introduces a new paradigm for knowledge dissemination and a foundation for the collaborative ecosystem of AI co-scientists.
Task-Agnostic Machine Learning-Assisted Inference
May 30, 2024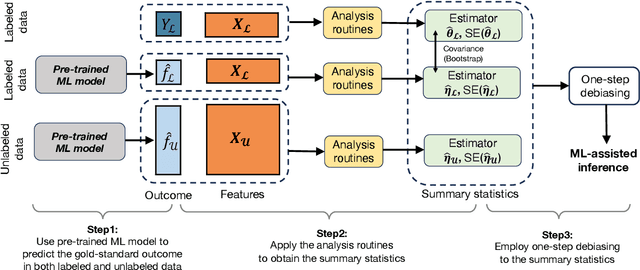
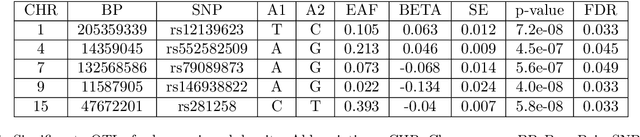
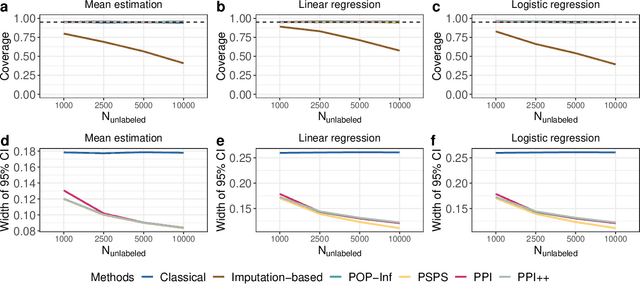
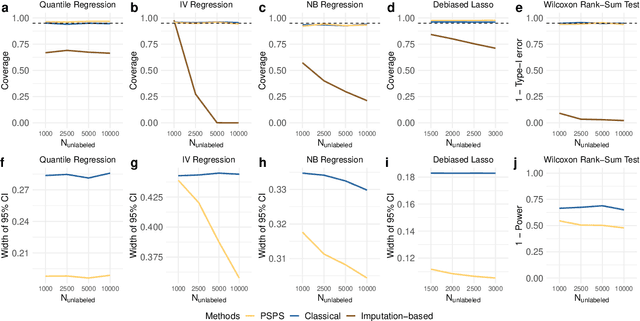
Abstract:Machine learning (ML) is playing an increasingly important role in scientific research. In conjunction with classical statistical approaches, ML-assisted analytical strategies have shown great promise in accelerating research findings. This has also opened up a whole new field of methodological research focusing on integrative approaches that leverage both ML and statistics to tackle data science challenges. One type of study that has quickly gained popularity employs ML to predict unobserved outcomes in massive samples and then uses the predicted outcomes in downstream statistical inference. However, existing methods designed to ensure the validity of this type of post-prediction inference are limited to very basic tasks such as linear regression analysis. This is because any extension of these approaches to new, more sophisticated statistical tasks requires task-specific algebraic derivations and software implementations, which ignores the massive library of existing software tools already developed for complex inference tasks and severely constrains the scope of post-prediction inference in real applications. To address this challenge, we propose a novel statistical framework for task-agnostic ML-assisted inference. It provides a post-prediction inference solution that can be easily plugged into almost any established data analysis routine. It delivers valid and efficient inference that is robust to arbitrary choices of ML models, while allowing nearly all existing analytical frameworks to be incorporated into the analysis of ML-predicted outcomes. Through extensive experiments, we showcase the validity, versatility, and superiority of our method compared to existing approaches.
Mask-adaptive Gated Convolution and Bi-directional Progressive Fusion Network for Depth Completion
Jan 15, 2024Abstract:Depth completion is a critical task for handling depth images with missing pixels, which can negatively impact further applications. Recent approaches have utilized Convolutional Neural Networks (CNNs) to reconstruct depth images with the assistance of color images. However, vanilla convolution has non-negligible drawbacks in handling missing pixels. To solve this problem, we propose a new model for depth completion based on an encoder-decoder structure. Our model introduces two key components: the Mask-adaptive Gated Convolution (MagaConv) architecture and the Bi-directional Progressive Fusion (BP-Fusion) module. The MagaConv architecture is designed to acquire precise depth features by modulating convolution operations with iteratively updated masks, while the BP-Fusion module progressively integrates depth and color features, utilizing consecutive bi-directional fusion structures in a global perspective. Extensive experiments on popular benchmarks, including NYU-Depth V2, DIML, and SUN RGB-D, demonstrate the superiority of our model over state-of-the-art methods. We achieved remarkable performance in completing depth maps and outperformed existing approaches in terms of accuracy and reliability.
Assumption-lean and Data-adaptive Post-Prediction Inference
Nov 23, 2023

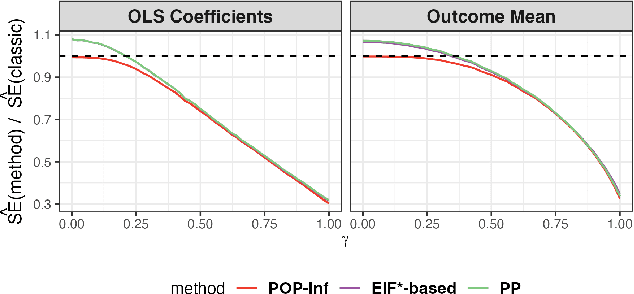

Abstract:A primary challenge facing modern scientific research is the limited availability of gold-standard data which can be both costly and labor-intensive to obtain. With the rapid development of machine learning (ML), scientists have relied on ML algorithms to predict these gold-standard outcomes with easily obtained covariates. However, these predicted outcomes are often used directly in subsequent statistical analyses, ignoring imprecision and heterogeneity introduced by the prediction procedure. This will likely result in false positive findings and invalid scientific conclusions. In this work, we introduce an assumption-lean and data-adaptive Post-Prediction Inference (POP-Inf) procedure that allows valid and powerful inference based on ML-predicted outcomes. Its "assumption-lean" property guarantees reliable statistical inference without assumptions on the ML-prediction, for a wide range of statistical quantities. Its "data-adaptive'" feature guarantees an efficiency gain over existing post-prediction inference methods, regardless of the accuracy of ML-prediction. We demonstrate the superiority and applicability of our method through simulations and large-scale genomic data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge