Jean-François Mangin
Neurospin, Gif-sur-Yvette, France
Ultra-high resolution multimodal MRI dense labelled holistic brain atlas
Jan 28, 2025



Abstract:In this paper, we introduce holiAtlas, a holistic, multimodal and high-resolution human brain atlas. This atlas covers different levels of details of the human brain anatomy, from the organ to the substructure level, using a new dense labelled protocol generated from the fusion of multiple local protocols at different scales. This atlas has been constructed averaging images and segmentations of 75 healthy subjects from the Human Connectome Project database. Specifically, MR images of T1, T2 and WMn (White Matter nulled) contrasts at 0.125 $mm^{3}$ resolution that were nonlinearly registered and averaged using symmetric group-wise normalisation to construct the atlas. At the finest level, the holiAtlas protocol has 350 different labels derived from 10 different delineation protocols. These labels were grouped at different scales to provide a holistic view of the brain at different levels in a coherent and consistent manner. This multiscale and multimodal atlas can be used for the development of new ultra-high resolution segmentation methods that can potentially leverage the early detection of neurological disorders.
Optimizing contrastive learning for cortical folding pattern detection
Jan 31, 2024Abstract:The human cerebral cortex has many bumps and grooves called gyri and sulci. Even though there is a high inter-individual consistency for the main cortical folds, this is not the case when we examine the exact shapes and details of the folding patterns. Because of this complexity, characterizing the cortical folding variability and relating them to subjects' behavioral characteristics or pathologies is still an open scientific problem. Classical approaches include labeling a few specific patterns, either manually or semi-automatically, based on geometric distances, but the recent availability of MRI image datasets of tens of thousands of subjects makes modern deep-learning techniques particularly attractive. Here, we build a self-supervised deep-learning model to detect folding patterns in the cingulate region. We train a contrastive self-supervised model (SimCLR) on both Human Connectome Project (1101 subjects) and UKBioBank (21070 subjects) datasets with topological-based augmentations on the cortical skeletons, which are topological objects that capture the shape of the folds. We explore several backbone architectures (convolutional network, DenseNet, and PointNet) for the SimCLR. For evaluation and testing, we perform a linear classification task on a database manually labeled for the presence of the "double-parallel" folding pattern in the cingulate region, which is related to schizophrenia characteristics. The best model, giving a test AUC of 0.76, is a convolutional network with 6 layers, a 10-dimensional latent space, a linear projection head, and using the branch-clipping augmentation. This is the first time that a self-supervised deep learning model has been applied to cortical skeletons on such a large dataset and quantitatively evaluated. We can now envisage the next step: applying it to other brain regions to detect other biomarkers.
GeoLab: Geometry-based Tractography Parcellation of Superficial White Matter
Mar 02, 2023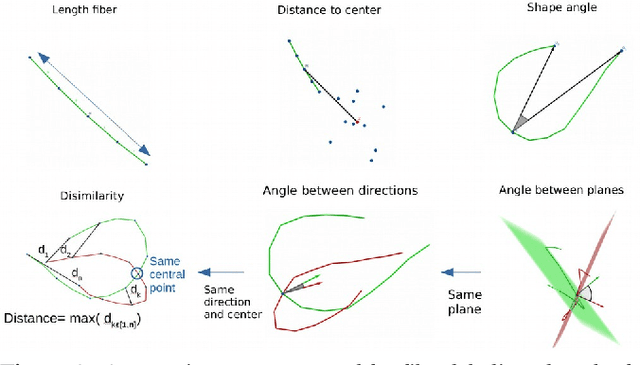
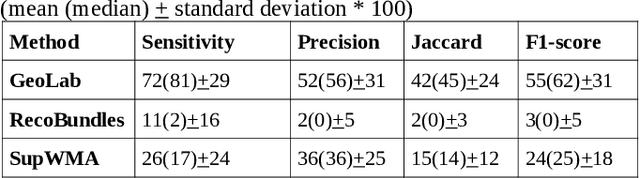
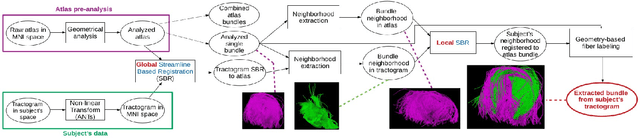
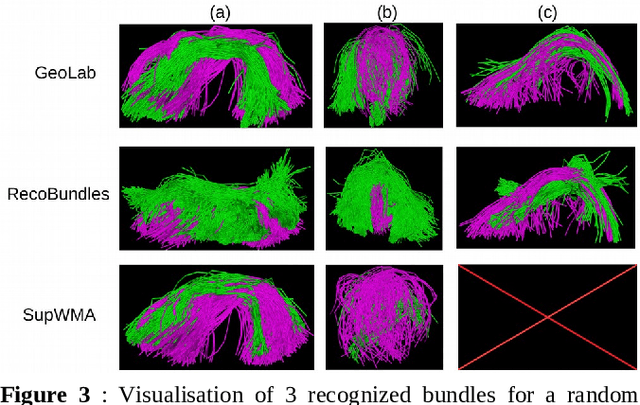
Abstract:Superficial white matter (SWM) has been less studied than long-range connections despite being of interest to clinical research, andfew tractography parcellation methods have been adapted to SWM. Here, we propose an efficient geometry-based parcellation method (GeoLab) that allows high-performance segmentation of hundreds of short white matter bundles from a subject. This method has been designed for the SWM atlas of EBRAINS European infrastructure, which is composed of 657 bundles. The atlas projection relies on the precomputed statistics of six bundle-specific geometrical properties of atlas streamlines. In the spirit of RecoBundles, a global and local streamline-based registration (SBR) is used to align the subject to the atlas space. Then, the streamlines are labeled taking into account the six geometrical parameters describing the similarity to the streamlines in the model bundle. Compared to other state-of-the-art methods, GeoLab allows the extraction of more bundles with a higher number of streamlines.
Identification of Rare Cortical Folding Patterns using Unsupervised Deep Learning
Nov 29, 2022Abstract:Like fingerprints, cortical folding patterns are unique to each brain even though they follow a general species-specific organization. Some folding patterns have been linked with neurodevelopmental disorders. However, due to the high inter-individual variability, the identification of rare folding patterns that could become biomarkers remains a very complex task. This paper proposes a novel unsupervised deep learning approach to identify rare folding patterns and assess the degree of deviations that can be detected. To this end, we preprocess the brain MR images to focus the learning on the folding morphology and train a beta-VAE to model the inter-individual variability of the folding. We compare the detection power of the latent space and of the reconstruction errors, using synthetic benchmarks and one actual rare configuration related to the central sulcus. Finally, we assess the generalization of our method on a developmental anomaly located in another region. Our results suggest that this method enables encoding relevant folding characteristics that can be enlightened and better interpreted based on the generative power of the beta-VAE. The latent space and the reconstruction errors bring complementary information and enable the identification of rare patterns of different nature. This method generalizes well to a different region on another dataset. Code is available at https://github.com/neurospin-projects/2022_lguillon_rare_folding_detection.
GeoSP: A parallel method for a cortical surface parcellation based on geodesic distance
Mar 26, 2021
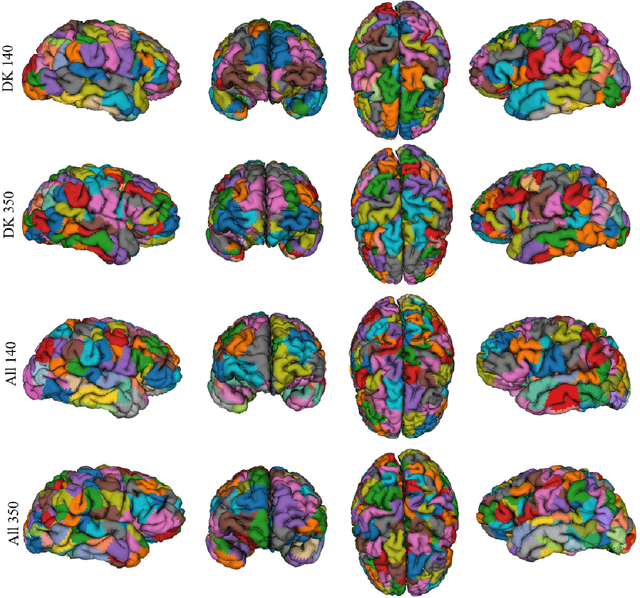
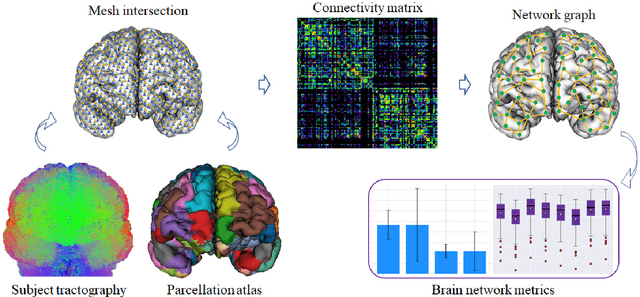
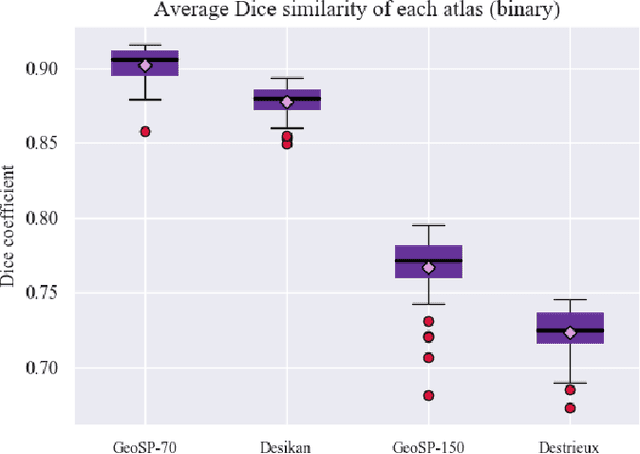
Abstract:We present GeoSP, a parallel method that creates a parcellation of the cortical mesh based on a geodesic distance, in order to consider gyri and sulci topology. The method represents the mesh with a graph and performs a K-means clustering in parallel. It has two modes of use, by default, it performs the geodesic cortical parcellation based on the boundaries of the anatomical parcels provided by the Desikan-Killiany atlas. The other mode performs the complete parcellation of the cortex. Results for both modes and with different values for the total number of sub-parcels show homogeneous sub-parcels. Furthermore, the execution time is 82 s for the whole cortex mode and 18 s for the Desikan-Killiany atlas subdivision, for a parcellation into 350 sub-parcels. The proposed method will be available to the community to perform the evaluation of data-driven cortical parcellations. As an example, we compared GeoSP parcellation with Desikan-Killiany and Destrieux atlases in 50 subjects, obtaining more homogeneous parcels for GeoSP and minor differences in structural connectivity reproducibility across subjects.
Cortical surface parcellation based on intra-subject white matter fiber clustering
Feb 16, 2020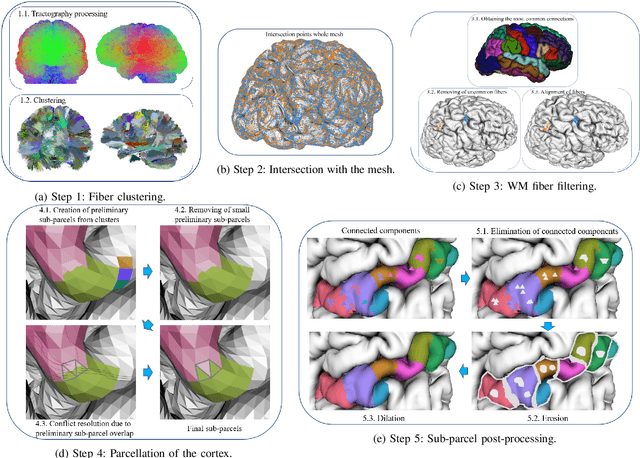
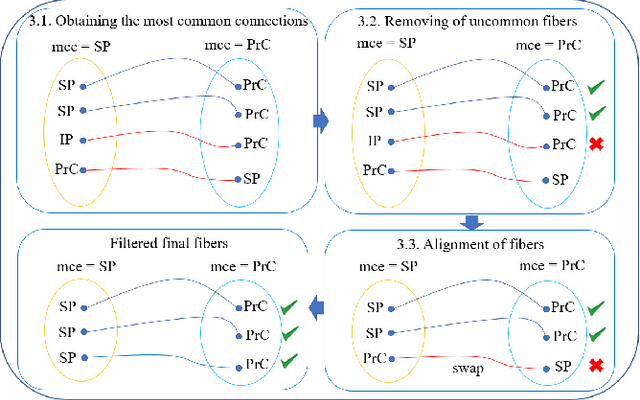
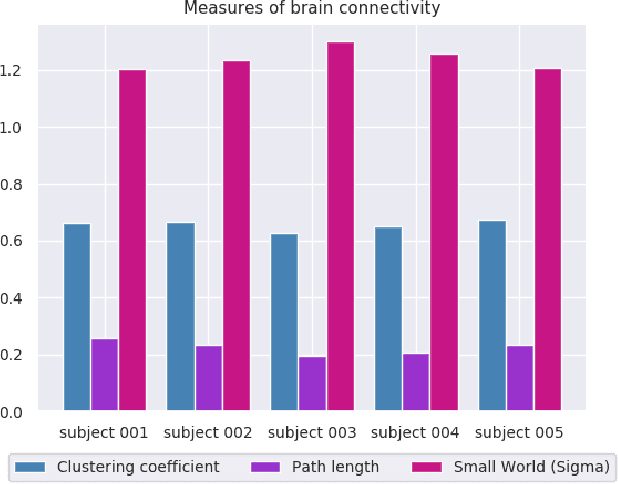
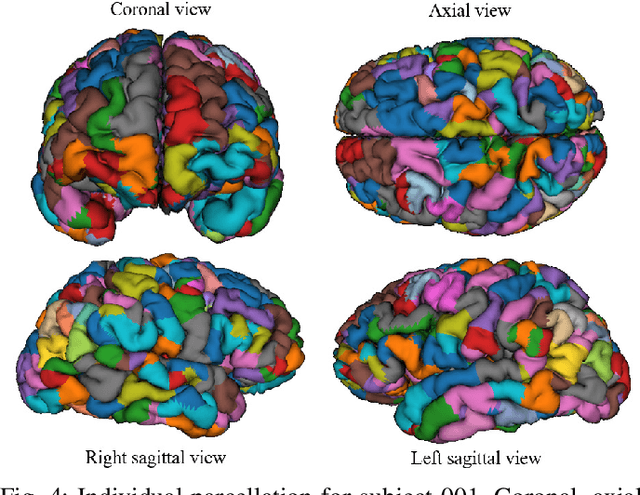
Abstract:We present a hybrid method that performs the complete parcellation of the cerebral cortex of an individual, based on the connectivity information of the white matter fibers from a whole-brain tractography dataset. The method consists of five steps, first intra-subject clustering is performed on the brain tractography. The fibers that make up each cluster are then intersected with the cortical mesh and then filtered to discard outliers. In addition, the method resolves the overlapping between the different intersection regions (sub-parcels) throughout the cortex efficiently. Finally, a post-processing is done to achieve more uniform sub-parcels. The output is the complete labeling of cortical mesh vertices, representing the different cortex sub-parcels, with strong connections to other sub-parcels. We evaluated our method with measures of brain connectivity such as functional segregation (clustering coefficient), functional integration (characteristic path length) and small-world. Results in five subjects from ARCHI database show a good individual cortical parcellation for each one, composed of about 200 subparcels per hemisphere and complying with these connectivity measures.
Parallel optimization of fiber bundle segmentation for massive tractography datasets
Dec 24, 2019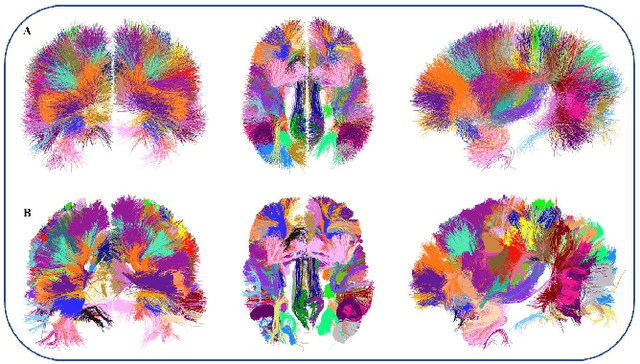
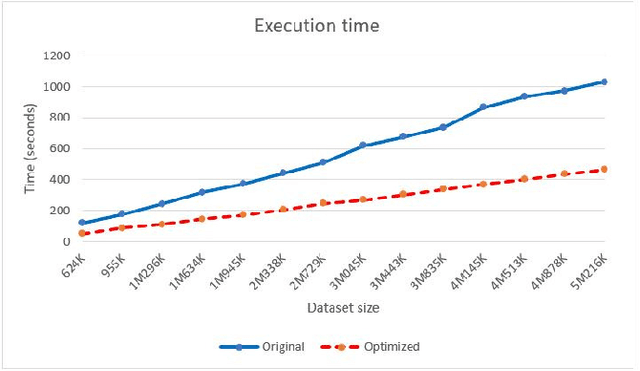
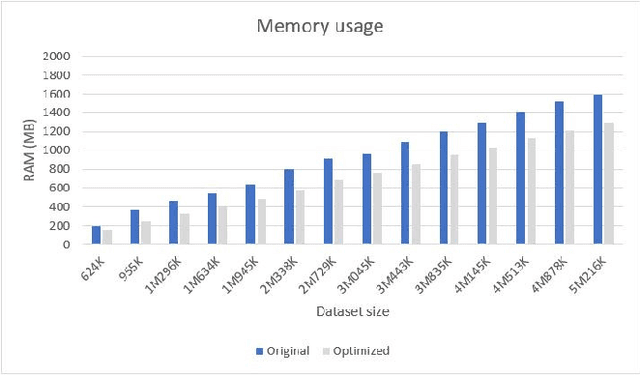
Abstract:We present an optimized algorithm that performs automatic classification of white matter fibers based on a multi-subject bundle atlas. We implemented a parallel algorithm that improves upon its previous version in both execution time and memory usage. Our new version uses the local memory of each processor, which leads to a reduction in execution time. Hence, it allows the analysis of bigger subject and/or atlas datasets. As a result, the segmentation of a subject of 4,145,000 fibers is reduced from about 14 minutes in the previous version to about 6 minutes, yielding an acceleration of 2.34. In addition, the new algorithm reduces the memory consumption of the previous version by a factor of 0.79.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge