Jacob Gildenblat
Preserving clusters and correlations: a dimensionality reduction method for exceptionally high global structure preservation
Mar 11, 2025Abstract:We present Preserving Clusters and Correlations (PCC), a novel dimensionality reduction (DR) method a novel dimensionality reduction (DR) method that achieves state-of-the-art global structure (GS) preservation while maintaining competitive local structure (LS) preservation. It optimizes two objectives: a GS preservation objective that preserves an approximation of Pearson and Spearman correlations between high- and low-dimensional distances, and an LS preservation objective that ensures clusters in the high-dimensional data are separable in the low-dimensional data. PCC has a state-of-the-art ability to preserve the GS while having competitive LS preservation. In addition, we show the correlation objective can be combined with UMAP to significantly improve its GS preservation with minimal degradation of the LS. We quantitatively benchmark PCC against existing methods and demonstrate its utility in medical imaging, and show PCC is a competitive DR technique that demonstrates superior GS preservation in our benchmarks.
Segmentation by Factorization: Unsupervised Semantic Segmentation for Pathology by Factorizing Foundation Model Features
Sep 09, 2024



Abstract:We introduce Segmentation by Factorization (F-SEG), an unsupervised segmentation method for pathology that generates segmentation masks from pre-trained deep learning models. F-SEG allows the use of pre-trained deep neural networks, including recently developed pathology foundation models, for semantic segmentation. It achieves this without requiring additional training or finetuning, by factorizing the spatial features extracted by the models into segmentation masks and their associated concept features. We create generic tissue phenotypes for H&E images by training clustering models for multiple numbers of clusters on features extracted from several deep learning models on The Cancer Genome Atlas Program (TCGA), and then show how the clusters can be used for factorizing corresponding segmentation masks using off-the-shelf deep learning models. Our results show that F-SEG provides robust unsupervised segmentation capabilities for H&E pathology images, and that the segmentation quality is greatly improved by utilizing pathology foundation models. We discuss and propose methods for evaluating the performance of unsupervised segmentation in pathology.
HEROHE Challenge: assessing HER2 status in breast cancer without immunohistochemistry or in situ hybridization
Nov 08, 2021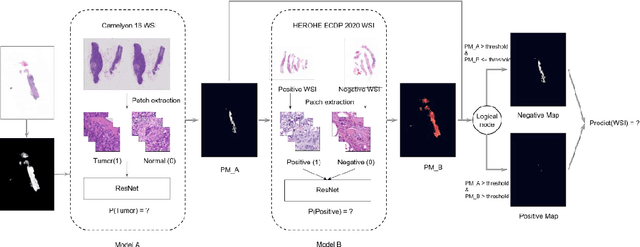
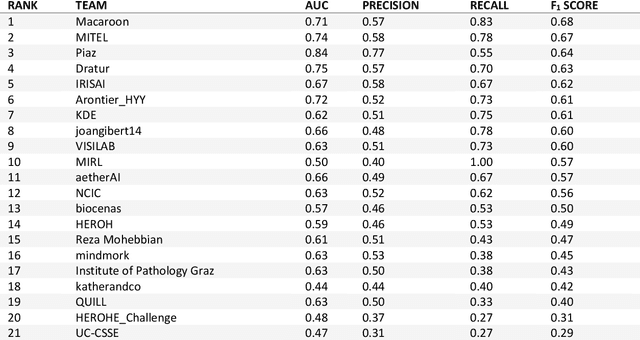
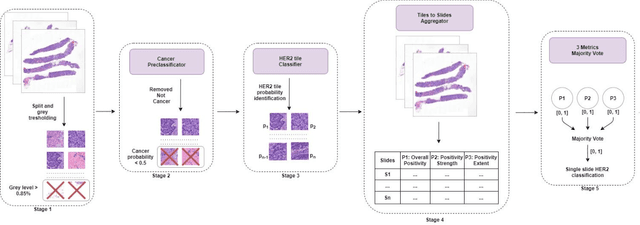
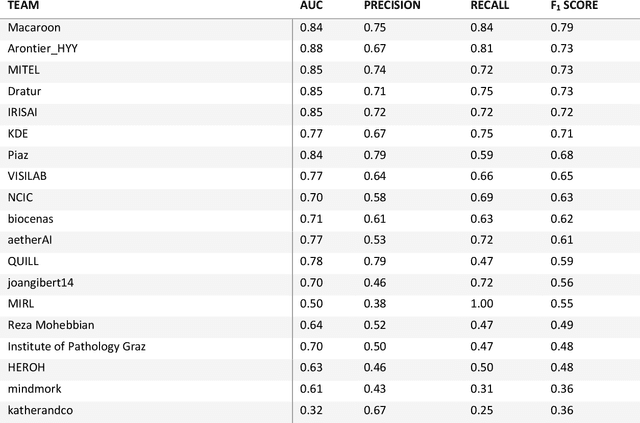
Abstract:Breast cancer is the most common malignancy in women, being responsible for more than half a million deaths every year. As such, early and accurate diagnosis is of paramount importance. Human expertise is required to diagnose and correctly classify breast cancer and define appropriate therapy, which depends on the evaluation of the expression of different biomarkers such as the transmembrane protein receptor HER2. This evaluation requires several steps, including special techniques such as immunohistochemistry or in situ hybridization to assess HER2 status. With the goal of reducing the number of steps and human bias in diagnosis, the HEROHE Challenge was organized, as a parallel event of the 16th European Congress on Digital Pathology, aiming to automate the assessment of the HER2 status based only on hematoxylin and eosin stained tissue sample of invasive breast cancer. Methods to assess HER2 status were presented by 21 teams worldwide and the results achieved by some of the proposed methods open potential perspectives to advance the state-of-the-art.
Certainty Pooling for Multiple Instance Learning
Aug 24, 2020
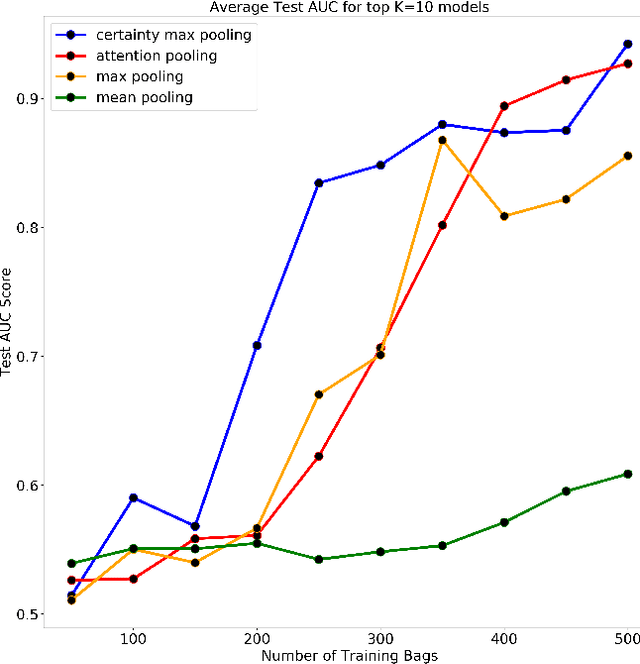


Abstract:Multiple Instance Learning is a form of weakly supervised learning in which the data is arranged in sets of instances called bags with one label assigned per bag. The bag level class prediction is derived from the multiple instances through application of a permutation invariant pooling operator on instance predictions or embeddings. We present a novel pooling operator called \textbf{Certainty Pooling} which incorporates the model certainty into bag predictions resulting in a more robust and explainable model. We compare our proposed method with other pooling operators in controlled experiments with low evidence ratio bags based on MNIST, as well as on a real life histopathology dataset - Camelyon16. Our method outperforms other methods in both bag level and instance level prediction, especially when only small training sets are available. We discuss the rationale behind our approach and the reasons for its superiority for these types of datasets.
Self-Supervised Similarity Learning for Digital Pathology
May 20, 2019



Abstract:Using features extracted from networks pretrained on ImageNet is a common practice in applications of deep learning for digital pathology. However it presents the downside of missing domain specific image information. In digital pathology, supervised training data is expensive and difficult to collect. We propose a self supervised method for feature extraction by similarity learning on whole slide images (WSI) that is simple to implement and allows creation of robust and compact image descriptors. We train a siamese network, exploiting image spatial continuity and assuming spatially adjacent tiles in the image are more similar to each other than distant tiles. Our network outputs feature vectors of length 128, which allows dramatically lower memory storage and faster processing than networks pretrained on ImageNet. We apply the method on digital pathology whole slide images (WSI) from the Camelyon16 train set and assess and compare our method by measuring image retrieval of tumor tiles and descriptor pair distance ratio for distant/near tiles in the Camelyon16 test set. We show that our method yields better retrieval task results than existing ImageNet based and generic self-supervised feature extraction methods. To the best of our knowledge, this is also the first published method for self supervised learning tailored for digital pathology.
Virtualization of tissue staining in digital pathology using an unsupervised deep learning approach
Oct 15, 2018



Abstract:Histopathological evaluation of tissue samples is a key practice in patient diagnosis and drug development, especially in oncology. Historically, Hematoxylin and Eosin (H&E) has been used by pathologists as a gold standard staining. However, in many cases, various target specific stains, including immunohistochemistry (IHC), are needed in order to highlight specific structures in the tissue. As tissue is scarce and staining procedures are tedious, it would be beneficial to generate images of stained tissue virtually. Virtual staining could also generate in-silico multiplexing of different stains on the same tissue segment. In this paper, we present a sample application that generates FAP-CK virtual IHC images from Ki67-CD8 real IHC images using an unsupervised deep learning approach based on CycleGAN. We also propose a method to deal with tiling artifacts caused by normalization layers and we validate our approach by comparing the results of tissue analysis algorithms for virtual and real images.
Generalizing multistain immunohistochemistry tissue segmentation using one-shot color deconvolution deep neural networks
Sep 22, 2018



Abstract:A key challenge in cancer immunotherapy biomarker research is quantification of pattern changes in microscopic whole slide images of tumor biopsies. Different cell types tend to migrate into various tissue compartments and form variable distribution patterns. Drug development requires correlative analysis of various biomarkers in and between the tissue compartments. To enable that, tissue slides are manually annotated by expert pathologists. Manual annotation of tissue slides is a labor intensive, tedious and error-prone task. Automation of this annotation process can improve accuracy and consistency while reducing workload and cost in a way that will positively influence drug development efforts. In this paper we present a novel one-shot color deconvolution deep learning method to automatically segment and annotate digitized slide images with multiple stainings into compartments of tumor, healthy tissue, and necrosis. We address the task in the context of drug development where multiple stains, tissue and tumor types exist and look into solutions for generalizations over these image populations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge