Jürgen Hetzel
Federated Learning in Multi-Center Critical Care Research: A Systematic Case Study using the eICU Database
Apr 20, 2022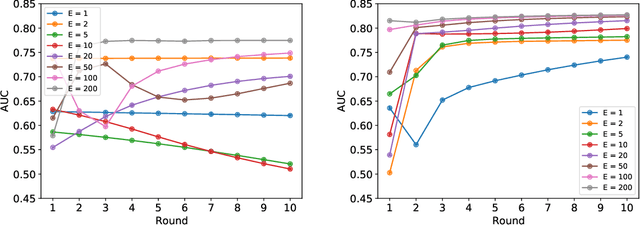
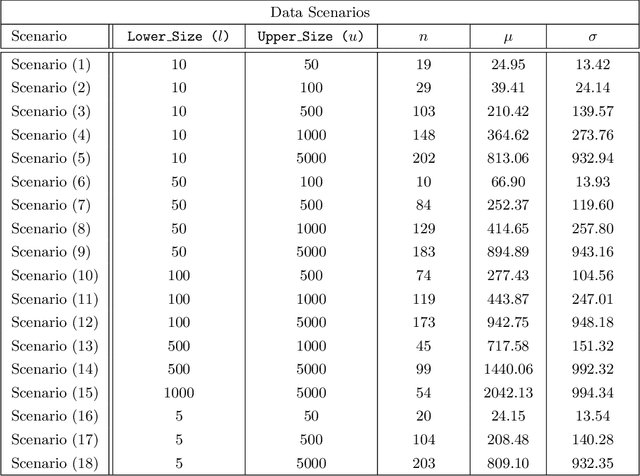
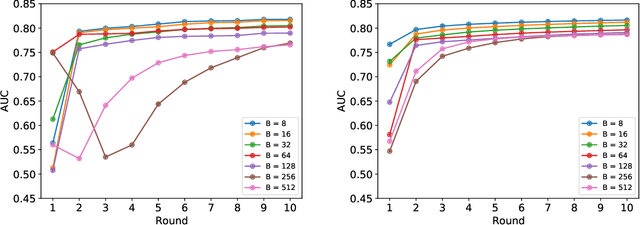
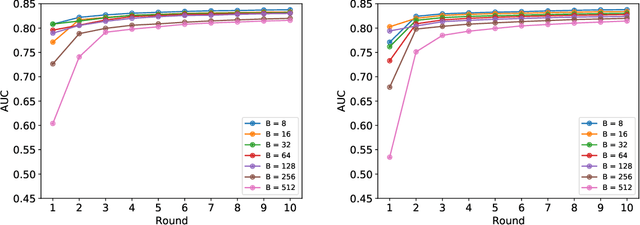
Abstract:Federated learning (FL) has been proposed as a method to train a model on different units without exchanging data. This offers great opportunities in the healthcare sector, where large datasets are available but cannot be shared to ensure patient privacy. We systematically investigate the effectiveness of FL on the publicly available eICU dataset for predicting the survival of each ICU stay. We employ Federated Averaging as the main practical algorithm for FL and show how its performance changes by altering three key hyper-parameters, taking into account that clients can significantly vary in size. We find that in many settings, a large number of local training epochs improves the performance while at the same time reducing communication costs. Furthermore, we outline in which settings it is possible to have only a low number of hospitals participating in each federated update round. When many hospitals with low patient counts are involved, the effect of overfitting can be avoided by decreasing the batchsize. This study thus contributes toward identifying suitable settings for running distributed algorithms such as FL on clinical datasets.
Overcoming Barriers to Data Sharing with Medical Image Generation: A Comprehensive Evaluation
Nov 29, 2020
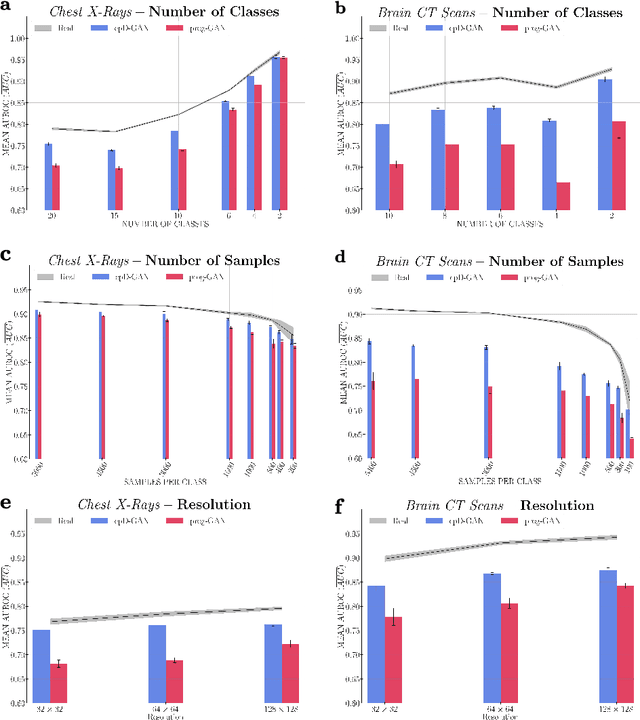
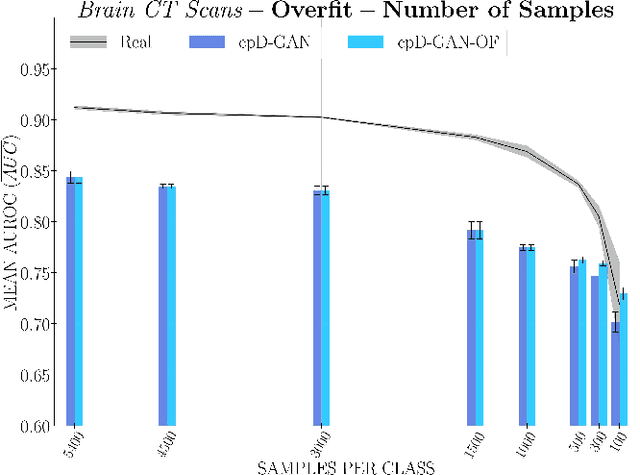
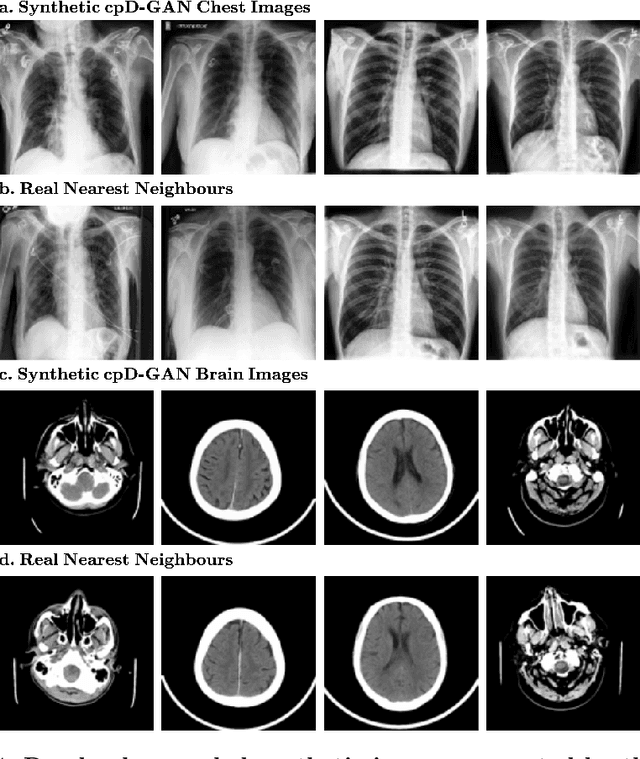
Abstract:Privacy concerns around sharing personally identifiable information are a major practical barrier to data sharing in medical research. However, in many cases, researchers have no interest in a particular individual's information but rather aim to derive insights at the level of cohorts. Here, we utilize Generative Adversarial Networks (GANs) to create derived medical imaging datasets consisting entirely of synthetic patient data. The synthetic images ideally have, in aggregate, similar statistical properties to those of a source dataset but do not contain sensitive personal information. We assess the quality of synthetic data generated by two GAN models for chest radiographs with 14 different radiology findings and brain computed tomography (CT) scans with six types of intracranial hemorrhages. We measure the synthetic image quality by the performance difference of predictive models trained on either the synthetic or the real dataset. We find that synthetic data performance disproportionately benefits from a reduced number of unique label combinations and determine at what number of samples per class overfitting effects start to dominate GAN training. Our open-source benchmark findings also indicate that synthetic data generation can benefit from higher levels of spatial resolution. We additionally conducted a reader study in which trained radiologists do not perform better than random on discriminating between synthetic and real medical images for both data modalities to a statistically significant extent. Our study offers valuable guidelines and outlines practical conditions under which insights derived from synthetic medical images are similar to those that would have been derived from real imaging data. Our results indicate that synthetic data sharing may be an attractive and privacy-preserving alternative to sharing real patient-level data in the right settings.
Real-time Prediction of COVID-19 related Mortality using Electronic Health Records
Aug 31, 2020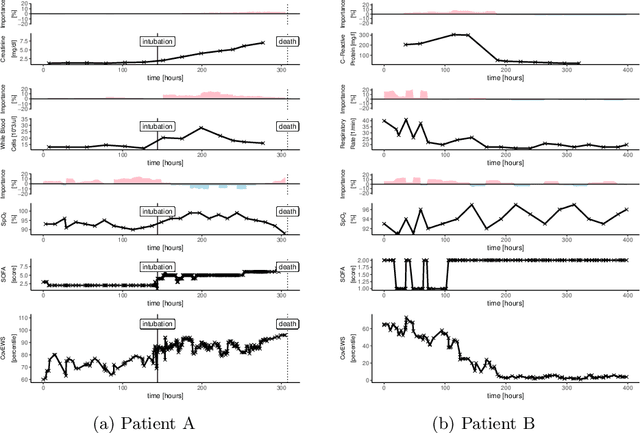
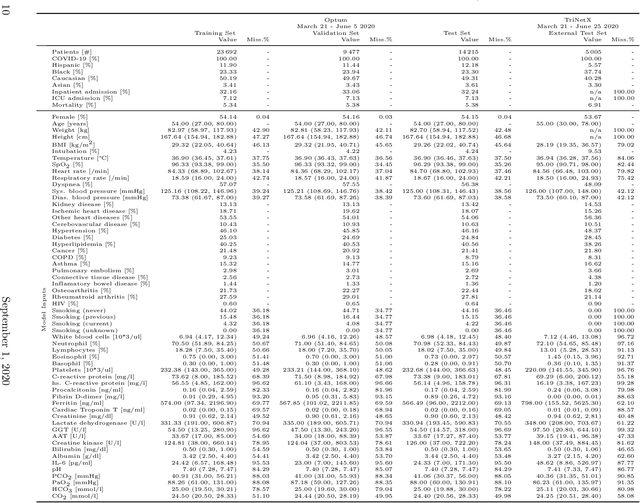
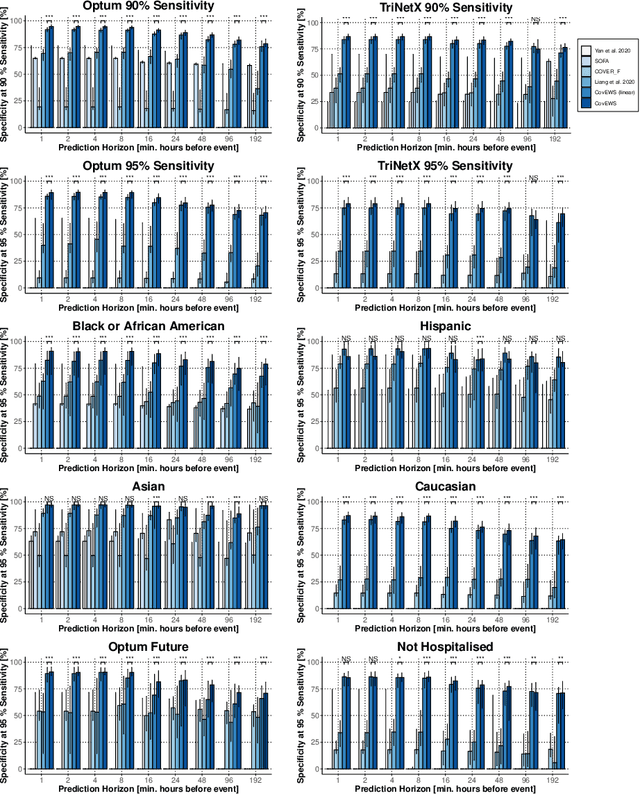
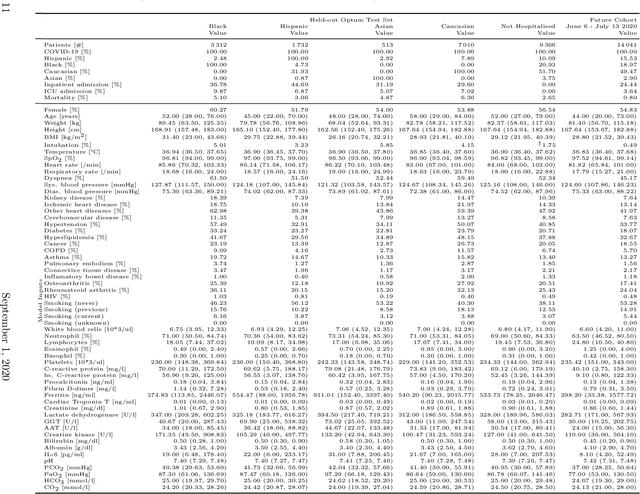
Abstract:Coronavirus Disease 2019 (COVID-19) is an emerging respiratory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with rapid human-to-human transmission and a high case fatality rate particularly in older patients. Due to the exponential growth of infections, many healthcare systems across the world are under pressure to care for increasing amounts of at-risk patients. Given the high number of infected patients, identifying patients with the highest mortality risk early is critical to enable effective intervention and optimal prioritisation of care. Here, we present the COVID-19 Early Warning System (CovEWS), a clinical risk scoring system for assessing COVID-19 related mortality risk. CovEWS provides continuous real-time risk scores for individual patients with clinically meaningful predictive performance up to 192 hours (8 days) in advance, and is automatically derived from patients' electronic health records (EHRs) using machine learning. We trained and evaluated CovEWS using de-identified data from a cohort of 66430 COVID-19 positive patients seen at over 69 healthcare institutions in the United States (US), Australia, Malaysia and India amounting to an aggregated total of over 2863 years of patient observation time. On an external test cohort of 5005 patients, CovEWS predicts COVID-19 related mortality from $78.8\%$ ($95\%$ confidence interval [CI]: $76.0$, $84.7\%$) to $69.4\%$ ($95\%$ CI: $57.6, 75.2\%$) specificity at a sensitivity greater than $95\%$ between respectively 1 and 192 hours prior to observed mortality events - significantly outperforming existing generic and COVID-19 specific clinical risk scores. CovEWS could enable clinicians to intervene at an earlier stage, and may therefore help in preventing or mitigating COVID-19 related mortality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge