August DuMont Schütte
Overcoming Barriers to Data Sharing with Medical Image Generation: A Comprehensive Evaluation
Nov 29, 2020
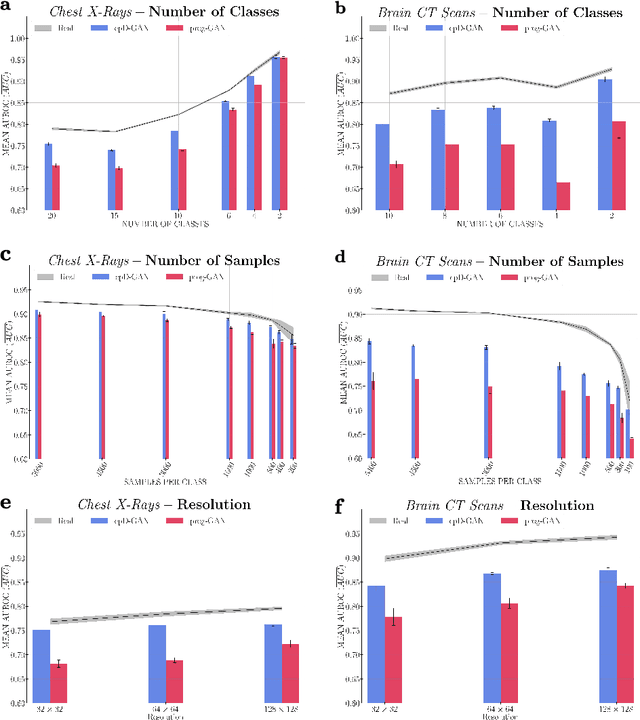
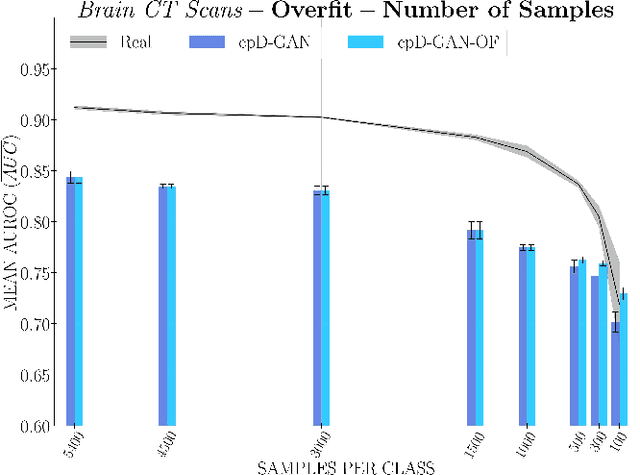
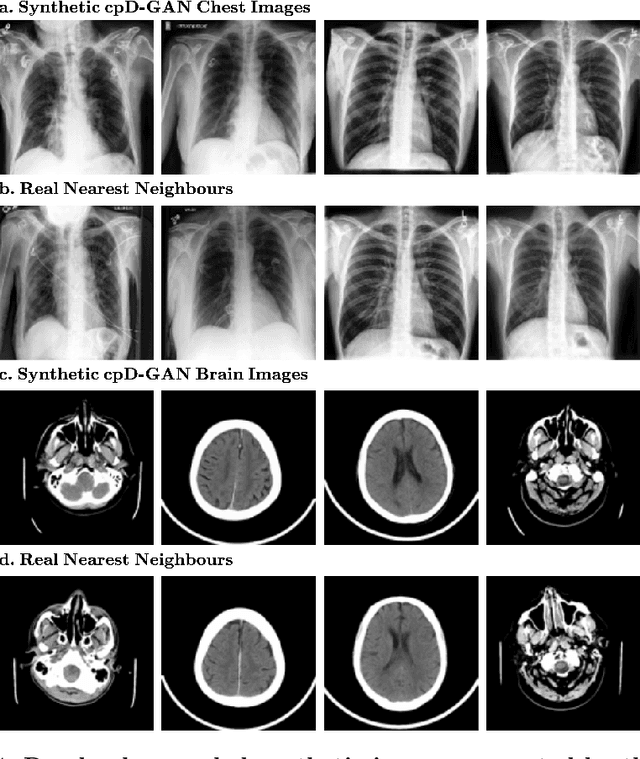
Abstract:Privacy concerns around sharing personally identifiable information are a major practical barrier to data sharing in medical research. However, in many cases, researchers have no interest in a particular individual's information but rather aim to derive insights at the level of cohorts. Here, we utilize Generative Adversarial Networks (GANs) to create derived medical imaging datasets consisting entirely of synthetic patient data. The synthetic images ideally have, in aggregate, similar statistical properties to those of a source dataset but do not contain sensitive personal information. We assess the quality of synthetic data generated by two GAN models for chest radiographs with 14 different radiology findings and brain computed tomography (CT) scans with six types of intracranial hemorrhages. We measure the synthetic image quality by the performance difference of predictive models trained on either the synthetic or the real dataset. We find that synthetic data performance disproportionately benefits from a reduced number of unique label combinations and determine at what number of samples per class overfitting effects start to dominate GAN training. Our open-source benchmark findings also indicate that synthetic data generation can benefit from higher levels of spatial resolution. We additionally conducted a reader study in which trained radiologists do not perform better than random on discriminating between synthetic and real medical images for both data modalities to a statistically significant extent. Our study offers valuable guidelines and outlines practical conditions under which insights derived from synthetic medical images are similar to those that would have been derived from real imaging data. Our results indicate that synthetic data sharing may be an attractive and privacy-preserving alternative to sharing real patient-level data in the right settings.
predCOVID-19: A Systematic Study of Clinical Predictive Models for Coronavirus Disease 2019
May 17, 2020



Abstract:Coronavirus Disease 2019 (COVID-19) is a rapidly emerging respiratory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Due to the rapid human-to-human transmission of SARS-CoV-2, many healthcare systems are at risk of exceeding their healthcare capacities, in particular in terms of SARS-CoV-2 tests, hospital and intensive care unit (ICU) beds and mechanical ventilators. Predictive algorithms could potentially ease the strain on healthcare systems by identifying those who are most likely to receive a positive SARS-CoV-2 test, be hospitalised or admitted to the ICU. Here, we study clinical predictive models that estimate, using machine learning and based on routinely collected clinical data, which patients are likely to receive a positive SARS-CoV-2 test, require hospitalisation or intensive care. To evaluate the predictive performance of our models, we perform a retrospective evaluation on clinical and blood analysis data from a cohort of 5644 patients. Our experimental results indicate that our predictive models identify (i) patients that test positive for SARS-CoV-2 a priori at a sensitivity of 75% (95% CI: 67%, 81%) and a specificity of 49% (95% CI: 46%, 51%), (ii) SARS-CoV-2 positive patients that require hospitalisation with 0.92 AUC (95% CI: 0.81, 0.98), and (iii) SARS-CoV-2 positive patients that require critical care with 0.98 AUC (95% CI: 0.95, 1.00). In addition, we determine which clinical features are predictive to what degree for each of the aforementioned clinical tasks. Our results indicate that predictive models trained on routinely collected clinical data could be used to predict clinical pathways for COVID-19, and therefore help inform care and prioritise resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge