Jörg K. Wegner
Modern Hopfield Networks for Few- and Zero-Shot Reaction Prediction
Apr 07, 2021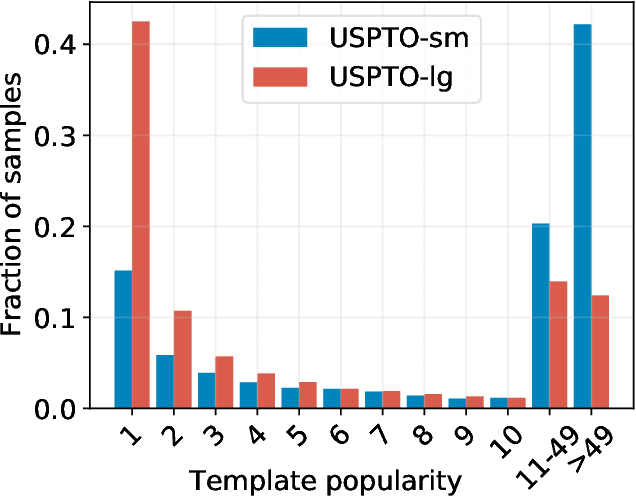
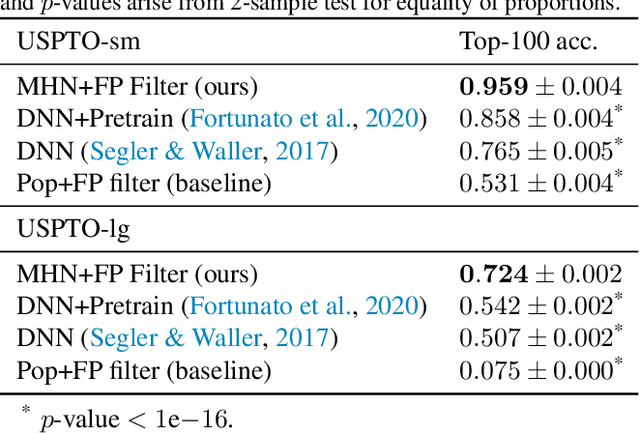
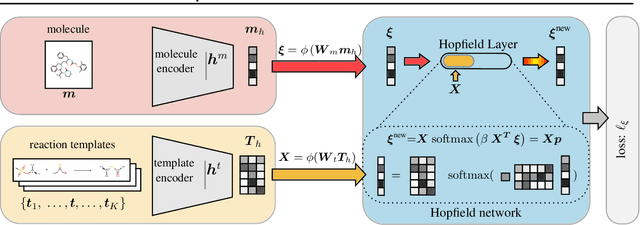
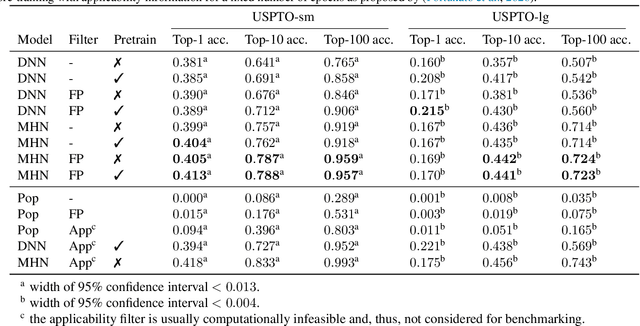
Abstract:An essential step in the discovery of new drugs and materials is the synthesis of a molecule that exists so far only as an idea to test its biological and physical properties. While computer-aided design of virtual molecules has made large progress, computer-assisted synthesis planning (CASP) to realize physical molecules is still in its infancy and lacks a performance level that would enable large-scale molecule discovery. CASP supports the search for multi-step synthesis routes, which is very challenging due to high branching factors in each synthesis step and the hidden rules that govern the reactions. The central and repeatedly applied step in CASP is reaction prediction, for which machine learning methods yield the best performance. We propose a novel reaction prediction approach that uses a deep learning architecture with modern Hopfield networks (MHNs) that is optimized by contrastive learning. An MHN is an associative memory that can store and retrieve chemical reactions in each layer of a deep learning architecture. We show that our MHN contrastive learning approach enables few- and zero-shot learning for reaction prediction which, in contrast to previous methods, can deal with rare, single, or even no training example(s) for a reaction. On a well established benchmark, our MHN approach pushes the state-of-the-art performance up by a large margin as it improves the predictive top-100 accuracy from $0.858\pm0.004$ to $0.959\pm0.004$. This advance might pave the way to large-scale molecule discovery.
Macau: Scalable Bayesian Multi-relational Factorization with Side Information using MCMC
Dec 17, 2015

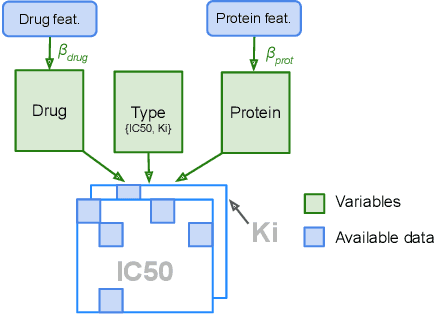
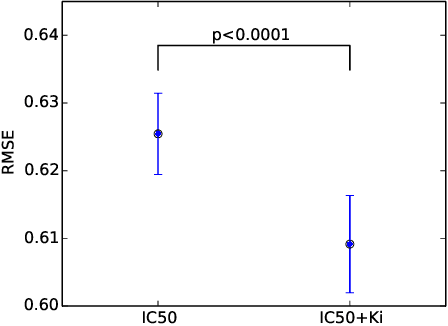
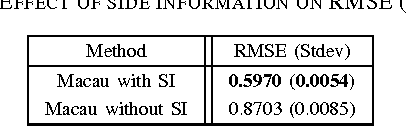
Abstract:We propose Macau, a powerful and flexible Bayesian factorization method for heterogeneous data. Our model can factorize any set of entities and relations that can be represented by a relational model, including tensors and also multiple relations for each entity. Macau can also incorporate side information, specifically entity and relation features, which are crucial for predicting sparsely observed relations. Macau scales to millions of entity instances, hundred millions of observations, and sparse entity features with millions of dimensions. To achieve the scale up, we specially designed sampling procedure for entity and relation features that relies primarily on noise injection in linear regressions. We show performance and advanced features of Macau in a set of experiments, including challenging drug-protein activity prediction task.
Highly Scalable Tensor Factorization for Prediction of Drug-Protein Interaction Type
Dec 01, 2015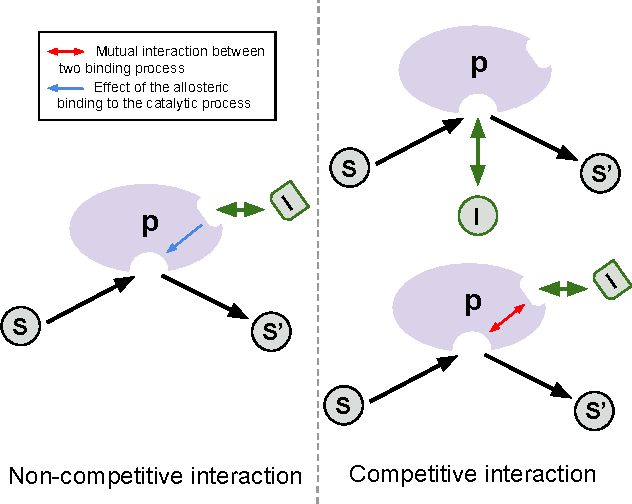
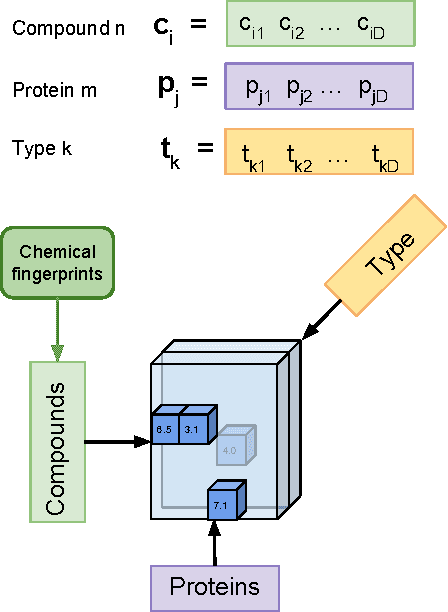
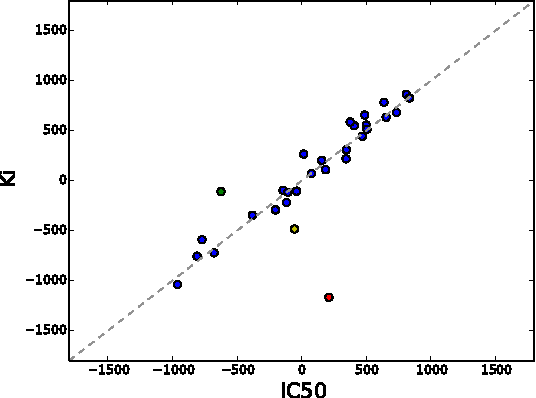
Abstract:The understanding of the type of inhibitory interaction plays an important role in drug design. Therefore, researchers are interested to know whether a drug has competitive or non-competitive interaction to particular protein targets. Method: to analyze the interaction types we propose factorization method Macau which allows us to combine different measurement types into a single tensor together with proteins and compounds. The compounds are characterized by high dimensional 2D ECFP fingerprints. The novelty of the proposed method is that using a specially designed noise injection MCMC sampler it can incorporate high dimensional side information, i.e., millions of unique 2D ECFP compound features, even for large scale datasets of millions of compounds. Without the side information, in this case, the tensor factorization would be practically futile. Results: using public IC50 and Ki data from ChEMBL we trained a model from where we can identify the latent subspace separating the two measurement types (IC50 and Ki). The results suggest the proposed method can detect the competitive inhibitory activity between compounds and proteins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge