Iyyakutti Iyappan Ganapathi
CLDTracker: A Comprehensive Language Description for Visual Tracking
May 29, 2025Abstract:VOT remains a fundamental yet challenging task in computer vision due to dynamic appearance changes, occlusions, and background clutter. Traditional trackers, relying primarily on visual cues, often struggle in such complex scenarios. Recent advancements in VLMs have shown promise in semantic understanding for tasks like open-vocabulary detection and image captioning, suggesting their potential for VOT. However, the direct application of VLMs to VOT is hindered by critical limitations: the absence of a rich and comprehensive textual representation that semantically captures the target object's nuances, limiting the effective use of language information; inefficient fusion mechanisms that fail to optimally integrate visual and textual features, preventing a holistic understanding of the target; and a lack of temporal modeling of the target's evolving appearance in the language domain, leading to a disconnect between the initial description and the object's subsequent visual changes. To bridge these gaps and unlock the full potential of VLMs for VOT, we propose CLDTracker, a novel Comprehensive Language Description framework for robust visual Tracking. Our tracker introduces a dual-branch architecture consisting of a textual and a visual branch. In the textual branch, we construct a rich bag of textual descriptions derived by harnessing the powerful VLMs such as CLIP and GPT-4V, enriched with semantic and contextual cues to address the lack of rich textual representation. Experiments on six standard VOT benchmarks demonstrate that CLDTracker achieves SOTA performance, validating the effectiveness of leveraging robust and temporally-adaptive vision-language representations for tracking. Code and models are publicly available at: https://github.com/HamadYA/CLDTracker
Multi-Resolution Pathology-Language Pre-training Model with Text-Guided Visual Representation
Apr 26, 2025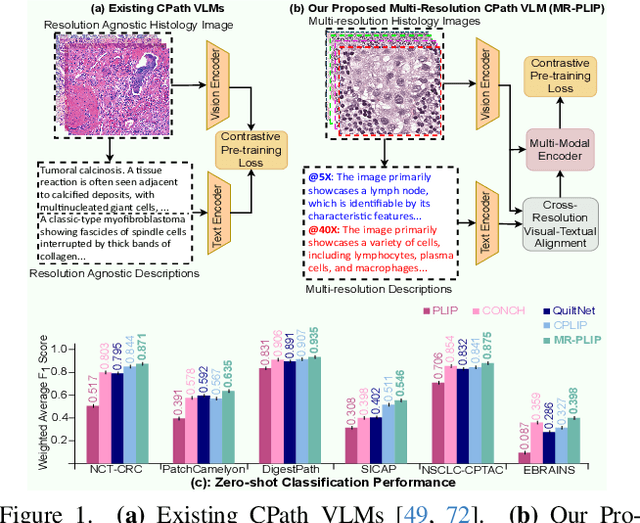
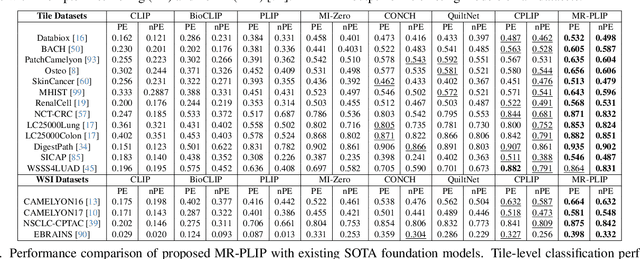
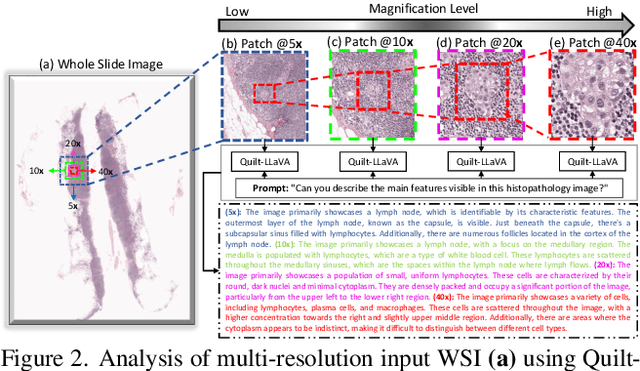
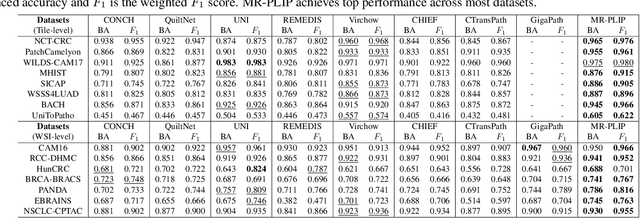
Abstract:In Computational Pathology (CPath), the introduction of Vision-Language Models (VLMs) has opened new avenues for research, focusing primarily on aligning image-text pairs at a single magnification level. However, this approach might not be sufficient for tasks like cancer subtype classification, tissue phenotyping, and survival analysis due to the limited level of detail that a single-resolution image can provide. Addressing this, we propose a novel multi-resolution paradigm leveraging Whole Slide Images (WSIs) to extract histology patches at multiple resolutions and generate corresponding textual descriptions through advanced CPath VLM. We introduce visual-textual alignment at multiple resolutions as well as cross-resolution alignment to establish more effective text-guided visual representations. Cross-resolution alignment using a multimodal encoder enhances the model's ability to capture context from multiple resolutions in histology images. Our model aims to capture a broader range of information, supported by novel loss functions, enriches feature representation, improves discriminative ability, and enhances generalization across different resolutions. Pre-trained on a comprehensive TCGA dataset with 34 million image-language pairs at various resolutions, our fine-tuned model outperforms state-of-the-art (SOTA) counterparts across multiple datasets and tasks, demonstrating its effectiveness in CPath. The code is available on GitHub at: https://github.com/BasitAlawode/MR-PLIP
DyCON: Dynamic Uncertainty-aware Consistency and Contrastive Learning for Semi-supervised Medical Image Segmentation
Apr 06, 2025Abstract:Semi-supervised learning in medical image segmentation leverages unlabeled data to reduce annotation burdens through consistency learning. However, current methods struggle with class imbalance and high uncertainty from pathology variations, leading to inaccurate segmentation in 3D medical images. To address these challenges, we present DyCON, a Dynamic Uncertainty-aware Consistency and Contrastive Learning framework that enhances the generalization of consistency methods with two complementary losses: Uncertainty-aware Consistency Loss (UnCL) and Focal Entropy-aware Contrastive Loss (FeCL). UnCL enforces global consistency by dynamically weighting the contribution of each voxel to the consistency loss based on its uncertainty, preserving high-uncertainty regions instead of filtering them out. Initially, UnCL prioritizes learning from uncertain voxels with lower penalties, encouraging the model to explore challenging regions. As training progress, the penalty shift towards confident voxels to refine predictions and ensure global consistency. Meanwhile, FeCL enhances local feature discrimination in imbalanced regions by introducing dual focal mechanisms and adaptive confidence adjustments into the contrastive principle. These mechanisms jointly prioritizes hard positives and negatives while focusing on uncertain sample pairs, effectively capturing subtle lesion variations under class imbalance. Extensive evaluations on four diverse medical image segmentation datasets (ISLES'22, BraTS'19, LA, Pancreas) show DyCON's superior performance against SOTA methods.
AquaticCLIP: A Vision-Language Foundation Model for Underwater Scene Analysis
Feb 03, 2025



Abstract:The preservation of aquatic biodiversity is critical in mitigating the effects of climate change. Aquatic scene understanding plays a pivotal role in aiding marine scientists in their decision-making processes. In this paper, we introduce AquaticCLIP, a novel contrastive language-image pre-training model tailored for aquatic scene understanding. AquaticCLIP presents a new unsupervised learning framework that aligns images and texts in aquatic environments, enabling tasks such as segmentation, classification, detection, and object counting. By leveraging our large-scale underwater image-text paired dataset without the need for ground-truth annotations, our model enriches existing vision-language models in the aquatic domain. For this purpose, we construct a 2 million underwater image-text paired dataset using heterogeneous resources, including YouTube, Netflix, NatGeo, etc. To fine-tune AquaticCLIP, we propose a prompt-guided vision encoder that progressively aggregates patch features via learnable prompts, while a vision-guided mechanism enhances the language encoder by incorporating visual context. The model is optimized through a contrastive pretraining loss to align visual and textual modalities. AquaticCLIP achieves notable performance improvements in zero-shot settings across multiple underwater computer vision tasks, outperforming existing methods in both robustness and interpretability. Our model sets a new benchmark for vision-language applications in underwater environments. The code and dataset for AquaticCLIP are publicly available on GitHub at xxx.
CPLIP: Zero-Shot Learning for Histopathology with Comprehensive Vision-Language Alignment
Jun 07, 2024



Abstract:This paper proposes Comprehensive Pathology Language Image Pre-training (CPLIP), a new unsupervised technique designed to enhance the alignment of images and text in histopathology for tasks such as classification and segmentation. This methodology enriches vision-language models by leveraging extensive data without needing ground truth annotations. CPLIP involves constructing a pathology-specific dictionary, generating textual descriptions for images using language models, and retrieving relevant images for each text snippet via a pre-trained model. The model is then fine-tuned using a many-to-many contrastive learning method to align complex interrelated concepts across both modalities. Evaluated across multiple histopathology tasks, CPLIP shows notable improvements in zero-shot learning scenarios, outperforming existing methods in both interpretability and robustness and setting a higher benchmark for the application of vision-language models in the field. To encourage further research and replication, the code for CPLIP is available on GitHub at https://cplip.github.io/
3D-TexSeg: Unsupervised Segmentation of 3D Texture using Mutual Transformer Learning
Nov 17, 2023



Abstract:Analysis of the 3D Texture is indispensable for various tasks, such as retrieval, segmentation, classification, and inspection of sculptures, knitted fabrics, and biological tissues. A 3D texture is a locally repeated surface variation independent of the surface's overall shape and can be determined using the local neighborhood and its characteristics. Existing techniques typically employ computer vision techniques that analyze a 3D mesh globally, derive features, and then utilize the obtained features for retrieval or classification. Several traditional and learning-based methods exist in the literature, however, only a few are on 3D texture, and nothing yet, to the best of our knowledge, on the unsupervised schemes. This paper presents an original framework for the unsupervised segmentation of the 3D texture on the mesh manifold. We approach this problem as binary surface segmentation, partitioning the mesh surface into textured and non-textured regions without prior annotation. We devise a mutual transformer-based system comprising a label generator and a cleaner. The two models take geometric image representations of the surface mesh facets and label them as texture or non-texture across an iterative mutual learning scheme. Extensive experiments on three publicly available datasets with diverse texture patterns demonstrate that the proposed framework outperforms standard and SOTA unsupervised techniques and competes reasonably with supervised methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge