Huafu Chen
LGLG-WPCA: An Effective Texture-based Method for Face Recognition
Nov 20, 2018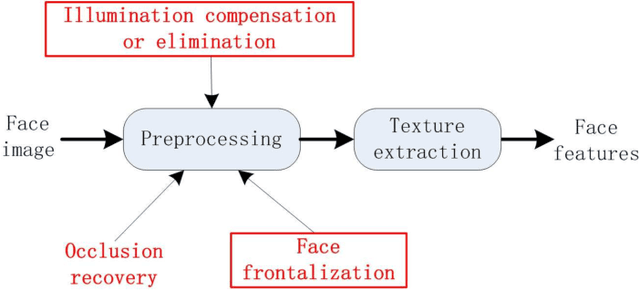
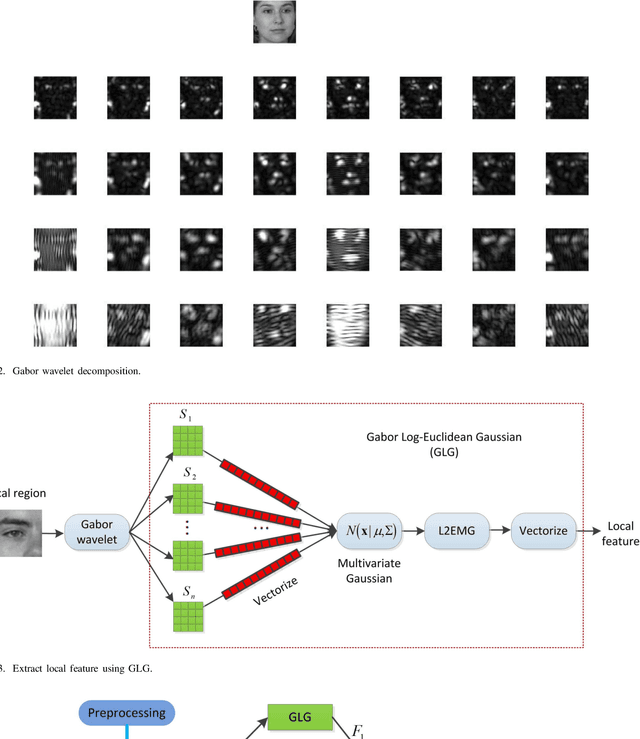
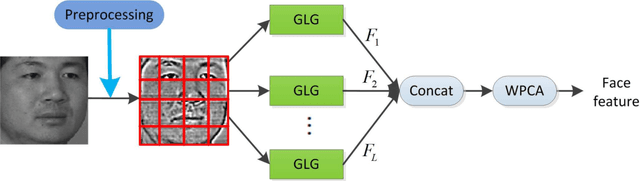
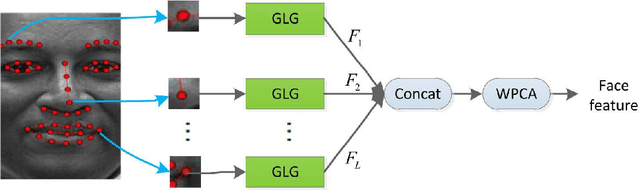
Abstract:This paper proposes an effective texture-based face feature extraction method which is based on Learning Gabor Log-Euclidean Gaussian, called LGLG-WPCA. LGLG-WPCA has the robust performance for adverse conditions such as varying poses, skin aging and uneven illumination. LGLG learns face features from the embedded multivariate Gaussian in Gabor wavelet domain using Whitening Principal Component Analysis (WPCA). In LGLG, we first employ Gabor wavelet to decompose the face, and then use the multivariate Gaussian distribution to fit Gabor subbands. Because the space of Gaussian is a Riemannian manifold and it is difficult to incorporate learning mechanism in the model. To address this issue, we use L$^2$EMG\cite{Li2017Local} to map the multidimensional Gaussian model to the linear space, and then use WPCA to learn facial features. Experiments show that our proposed method is an effective and promising face texture feature extraction technique.
A Novel Approach for Stable Selection of Informative Redundant Features from High Dimensional fMRI Data
May 25, 2016
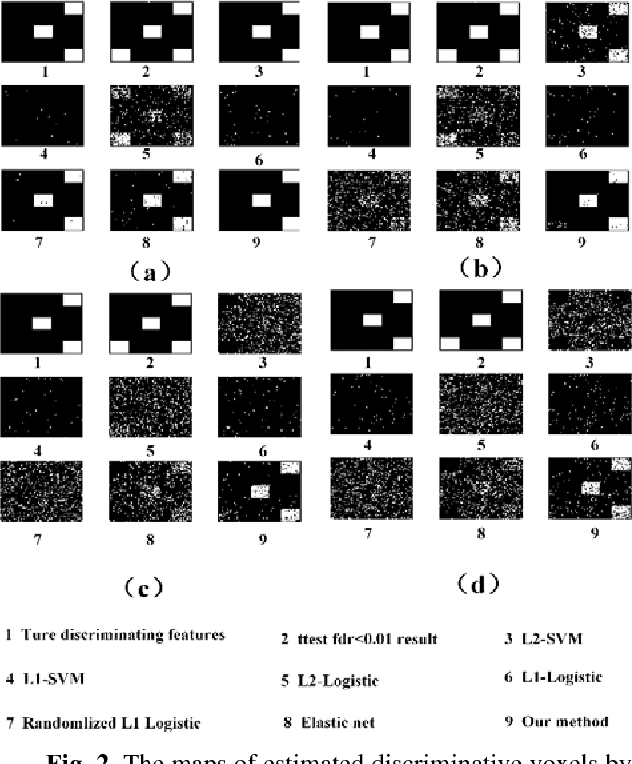
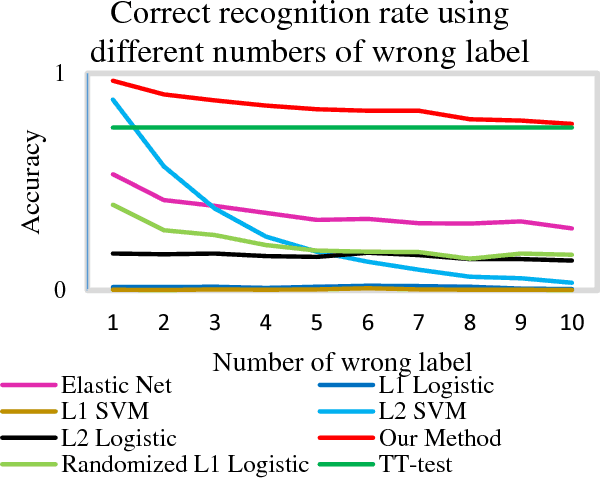
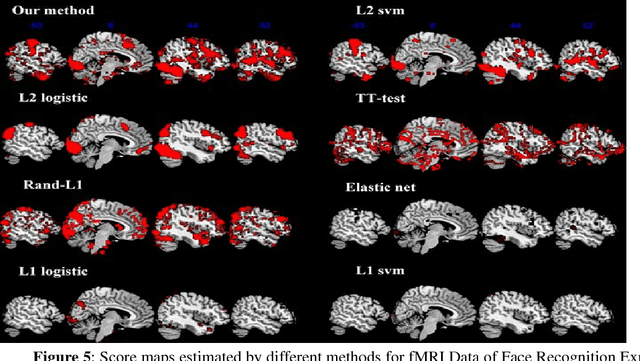
Abstract:Feature selection is among the most important components because it not only helps enhance the classification accuracy, but also or even more important provides potential biomarker discovery. However, traditional multivariate methods is likely to obtain unstable and unreliable results in case of an extremely high dimensional feature space and very limited training samples, where the features are often correlated or redundant. In order to improve the stability, generalization and interpretations of the discovered potential biomarker and enhance the robustness of the resultant classifier, the redundant but informative features need to be also selected. Therefore we introduced a novel feature selection method which combines a recent implementation of the stability selection approach and the elastic net approach. The advantage in terms of better control of false discoveries and missed discoveries of our approach, and the resulted better interpretability of the obtained potential biomarker is verified in both synthetic and real fMRI experiments. In addition, we are among the first to demonstrate the robustness of feature selection benefiting from the incorporation of stability selection and also among the first to demonstrate the possible unrobustness of the classical univariate two-sample t-test method. Specifically, we show the robustness of our feature selection results in existence of noisy (wrong) training labels, as well as the robustness of the resulted classifier based on our feature selection results in the existence of data variation, demonstrated by a multi-center attention-deficit/hyperactivity disorder (ADHD) fMRI data.
Randomized Structural Sparsity based Support Identification with Applications to Locating Activated or Discriminative Brain Areas: A Multi-center Reproducibility Study
Jun 07, 2015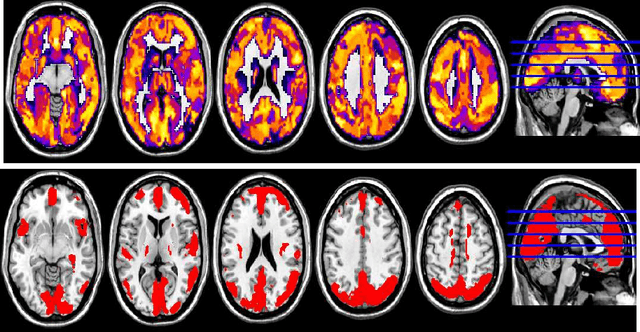
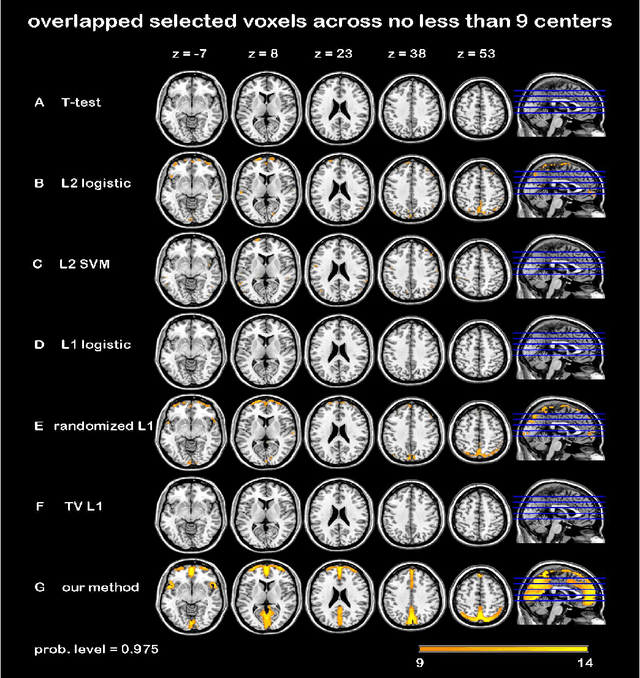
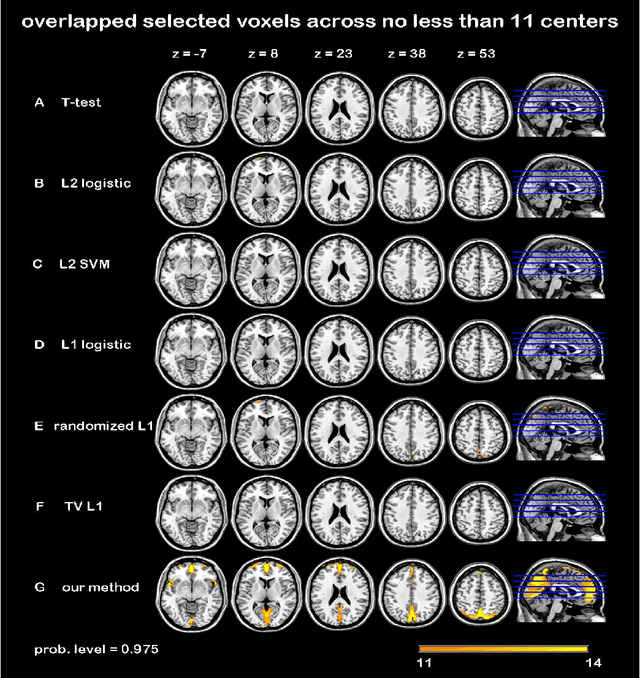

Abstract:In this paper, we focus on how to locate the relevant or discriminative brain regions related with external stimulus or certain mental decease, which is also called support identification, based on the neuroimaging data. The main difficulty lies in the extremely high dimensional voxel space and relatively few training samples, easily resulting in an unstable brain region discovery (or called feature selection in context of pattern recognition). When the training samples are from different centers and have betweencenter variations, it will be even harder to obtain a reliable and consistent result. Corresponding, we revisit our recently proposed algorithm based on stability selection and structural sparsity. It is applied to the multi-center MRI data analysis for the first time. A consistent and stable result is achieved across different centers despite the between-center data variation while many other state-of-the-art methods such as two sample t-test fail. Moreover, we have empirically showed that the performance of this algorithm is robust and insensitive to several of its key parameters. In addition, the support identification results on both functional MRI and structural MRI are interpretable and can be the potential biomarkers.
Randomized Structural Sparsity via Constrained Block Subsampling for Improved Sensitivity of Discriminative Voxel Identification
Jun 07, 2015



Abstract:In this paper, we consider voxel selection for functional Magnetic Resonance Imaging (fMRI) brain data with the aim of finding a more complete set of probably correlated discriminative voxels, thus improving interpretation of the discovered potential biomarkers. The main difficulty in doing this is an extremely high dimensional voxel space and few training samples, resulting in unreliable feature selection. In order to deal with the difficulty, stability selection has received a great deal of attention lately, especially due to its finite sample control of false discoveries and transparent principle for choosing a proper amount of regularization. However, it fails to make explicit use of the correlation property or structural information of these discriminative features and leads to large false negative rates. In other words, many relevant but probably correlated discriminative voxels are missed. Thus, we propose a new variant on stability selection "randomized structural sparsity", which incorporates the idea of structural sparsity. Numerical experiments demonstrate that our method can be superior in controlling for false negatives while also keeping the control of false positives inherited from stability selection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge