Xiaona Wang
LPGen: Enhancing High-Fidelity Landscape Painting Generation through Diffusion Model
Jul 25, 2024Abstract:Generating landscape paintings expands the possibilities of artistic creativity and imagination. Traditional landscape painting methods involve using ink or colored ink on rice paper, which requires substantial time and effort. These methods are susceptible to errors and inconsistencies and lack precise control over lines and colors. This paper presents LPGen, a high-fidelity, controllable model for landscape painting generation, introducing a novel multi-modal framework that integrates image prompts into the diffusion model. We extract its edges and contours by computing canny edges from the target landscape image. These, along with natural language text prompts and drawing style references, are fed into the latent diffusion model as conditions. We implement a decoupled cross-attention strategy to ensure compatibility between image and text prompts, facilitating multi-modal image generation. A decoder generates the final image. Quantitative and qualitative analyses demonstrate that our method outperforms existing approaches in landscape painting generation and exceeds the current state-of-the-art. The LPGen network effectively controls the composition and color of landscape paintings, generates more accurate images, and supports further research in deep learning-based landscape painting generation.
A Novel Approach for Stable Selection of Informative Redundant Features from High Dimensional fMRI Data
May 25, 2016
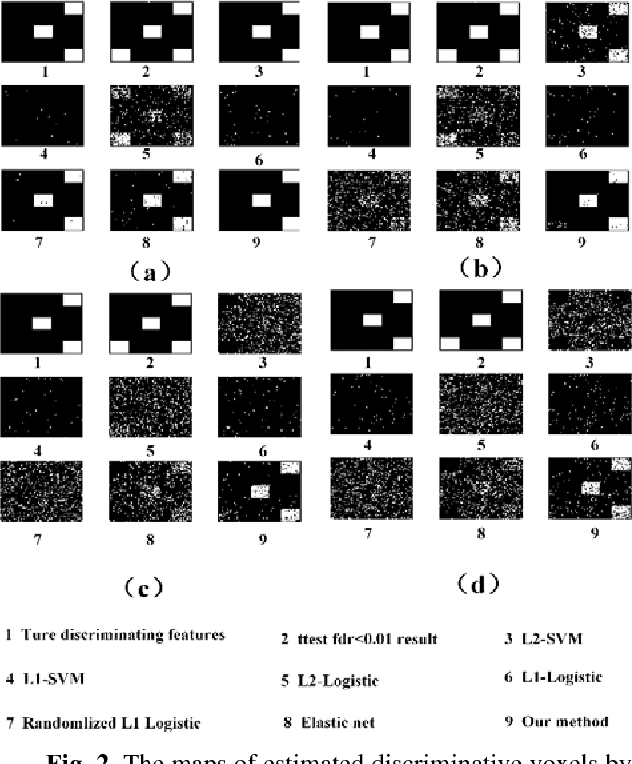
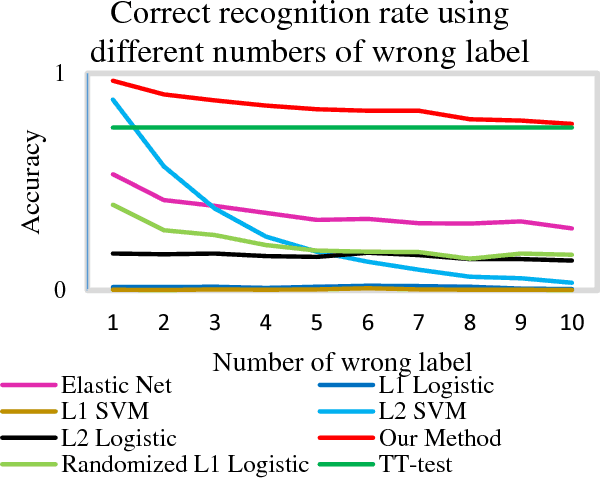
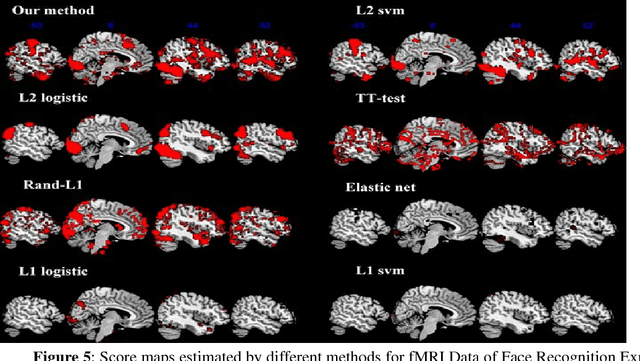
Abstract:Feature selection is among the most important components because it not only helps enhance the classification accuracy, but also or even more important provides potential biomarker discovery. However, traditional multivariate methods is likely to obtain unstable and unreliable results in case of an extremely high dimensional feature space and very limited training samples, where the features are often correlated or redundant. In order to improve the stability, generalization and interpretations of the discovered potential biomarker and enhance the robustness of the resultant classifier, the redundant but informative features need to be also selected. Therefore we introduced a novel feature selection method which combines a recent implementation of the stability selection approach and the elastic net approach. The advantage in terms of better control of false discoveries and missed discoveries of our approach, and the resulted better interpretability of the obtained potential biomarker is verified in both synthetic and real fMRI experiments. In addition, we are among the first to demonstrate the robustness of feature selection benefiting from the incorporation of stability selection and also among the first to demonstrate the possible unrobustness of the classical univariate two-sample t-test method. Specifically, we show the robustness of our feature selection results in existence of noisy (wrong) training labels, as well as the robustness of the resulted classifier based on our feature selection results in the existence of data variation, demonstrated by a multi-center attention-deficit/hyperactivity disorder (ADHD) fMRI data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge