Hua Su
Global-Local Detail Guided Transformer for Sea Ice Recognition in Optical Remote Sensing Images
May 21, 2024



Abstract:The recognition of sea ice is of great significance for reflecting climate change and ensuring the safety of ship navigation. Recently, many deep learning based methods have been proposed and applied to segment and recognize sea ice regions. However, the diverse scales of sea ice areas, the zigzag and fine edge contours, and the difficulty in distinguishing different types of sea ice pose challenges to existing sea ice recognition models. In this paper, a Global-Local Detail Guided Transformer (GDGT) method is proposed for sea ice recognition in optical remote sensing images. In GDGT, a global-local feature fusiont mechanism is designed to fuse global structural correlation features and local spatial detail features. Furthermore, a detail-guided decoder is developed to retain more high-resolution detail information during feature reconstruction for improving the performance of sea ice recognition. Experiments on the produced sea ice dataset demonstrated the effectiveness and advancement of GDGT.
* 5 pages, 5 figures
Global Mapping of Gene/Protein Interactions in PubMed Abstracts: A Framework and an Experiment with P53 Interactions
Apr 22, 2022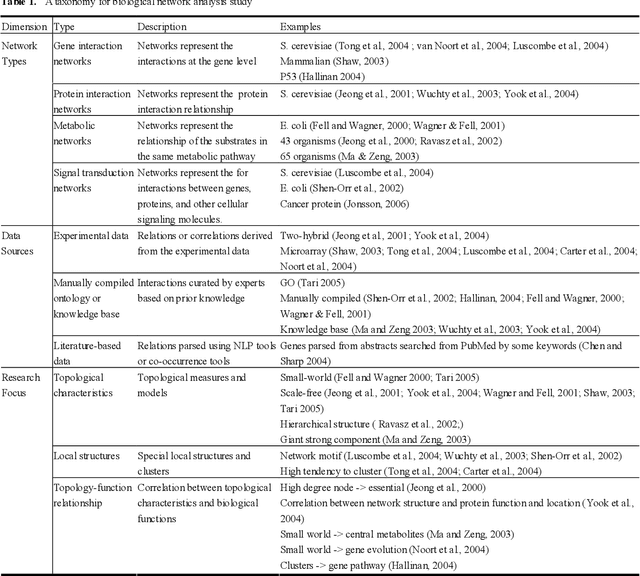
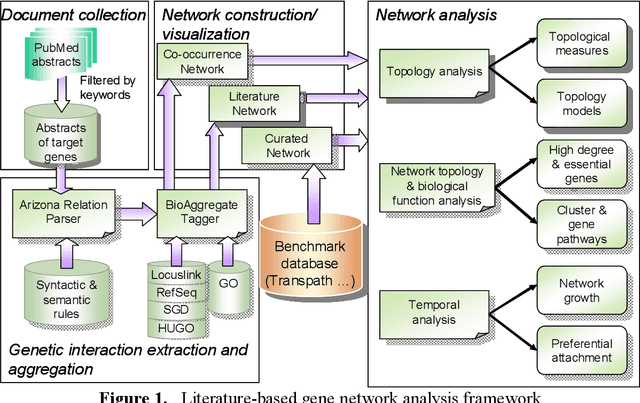
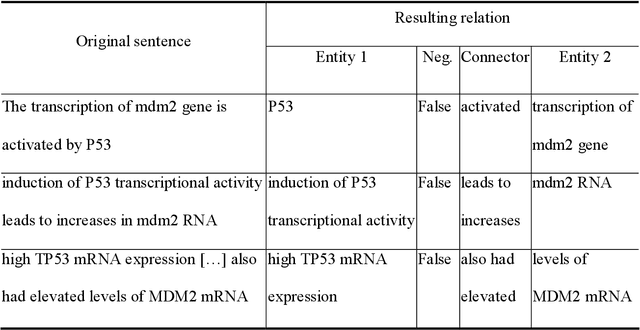
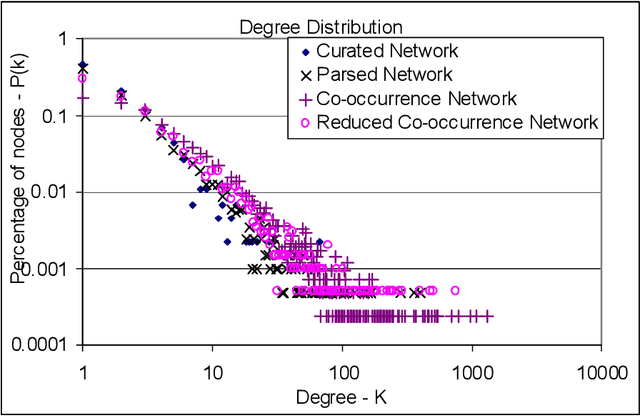
Abstract:Gene/protein interactions provide critical information for a thorough understanding of cellular processes. Recently, considerable interest and effort has been focused on the construction and analysis of genome-wide gene networks. The large body of biomedical literature is an important source of gene/protein interaction information. Recent advances in text mining tools have made it possible to automatically extract such documented interactions from free-text literature. In this paper, we propose a comprehensive framework for constructing and analyzing large-scale gene functional networks based on the gene/protein interactions extracted from biomedical literature repositories using text mining tools. Our proposed framework consists of analyses of the network topology, network topology-gene function relationship, and temporal network evolution to distill valuable information embedded in the gene functional interactions in literature. We demonstrate the application of the proposed framework using a testbed of P53-related PubMed abstracts, which shows that literature-based P53 networks exhibit small-world and scale-free properties. We also found that high degree genes in the literature-based networks have a high probability of appearing in the manually curated database and genes in the same pathway tend to form local clusters in our literature-based networks. Temporal analysis showed that genes interacting with many other genes tend to be involved in a large number of newly discovered interactions.
Learning Deep Implicit Functions for 3D Shapes with Dynamic Code Clouds
Mar 30, 2022



Abstract:Deep Implicit Function (DIF) has gained popularity as an efficient 3D shape representation. To capture geometry details, current methods usually learn DIF using local latent codes, which discretize the space into a regular 3D grid (or octree) and store local codes in grid points (or octree nodes). Given a query point, the local feature is computed by interpolating its neighboring local codes with their positions. However, the local codes are constrained at discrete and regular positions like grid points, which makes the code positions difficult to be optimized and limits their representation ability. To solve this problem, we propose to learn DIF with Dynamic Code Cloud, named DCC-DIF. Our method explicitly associates local codes with learnable position vectors, and the position vectors are continuous and can be dynamically optimized, which improves the representation ability. In addition, we propose a novel code position loss to optimize the code positions, which heuristically guides more local codes to be distributed around complex geometric details. In contrast to previous methods, our DCC-DIF represents 3D shapes more efficiently with a small amount of local codes, and improves the reconstruction quality. Experiments demonstrate that DCC-DIF achieves better performance over previous methods. Code and data are available at https://github.com/lity20/DCCDIF.
Global optimization of expensive black-box models based on asynchronous hybrid-criterion with interval reduction
Nov 29, 2018



Abstract:In this paper, a new sequential surrogate-based optimization (SSBO) algorithm is developed, which aims to improve the global search ability and local search efficiency for the global optimization of expensive black-box models. The proposed method involves three basic sub-criteria to infill new samples asynchronously to balance the global exploration and local exploitation. First, to capture the promising possible global optimal region, searching for the global optimum with genetic algorithm (GA) based on the current surrogate models of the objective and constraint functions. Second, to infill samples in the region with sparse samples to improve the global accuracy of the surrogate models, a grid searching with Latin hypercube sampling (LHS) with the current surrogate model is adopted to explore the sample space. Third, to accelerate the local searching efficiency, searching for a local optimum with sequential quadratic programming (SQP) based on the local surrogate models in the reduced interval, which involves some samples near the current optimum. When the new sample is too close to the existing ones, the new sample should be abandoned, due to the poor additional information. According to the three sub-criteria, the new samples are placed in the regions which have not been fully explored and includes the possible global optimum point. When a possible global optimum point is found, the local searching sub-criterion captures the local optimum around it rapidly. Numerical and engineering examples are used to verify the efficiency of the proposed method. The statistical results show that the proposed method has good global searching ability and efficiency.
Classification of Protein Crystallization X-Ray Images Using Major Convolutional Neural Network Architectures
May 11, 2018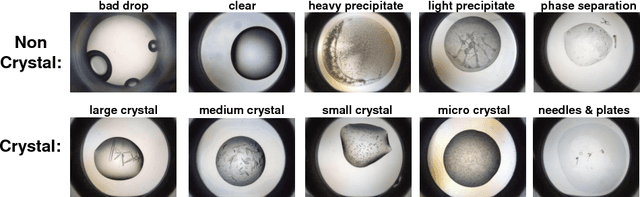
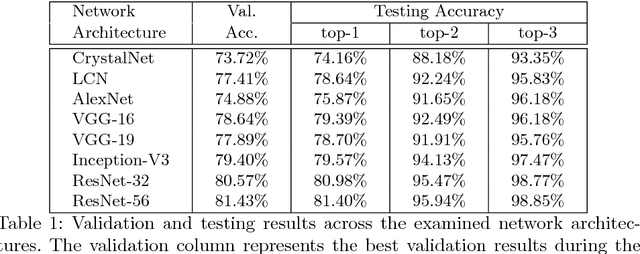
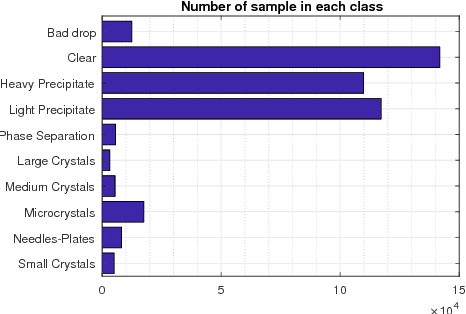
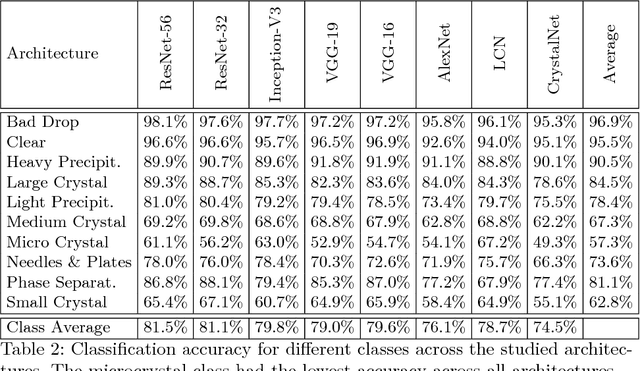
Abstract:The generation of protein crystals is necessary for the study of protein molecular function and structure. This is done empirically by processing large numbers of crystallization trials and inspecting them regularly in search of those with forming crystals. To avoid missing the hard-gained crystals, this visual inspection of the trial X-ray images is done manually as opposed to the existing less accurate machine learning methods. To achieve higher accuracy for automation, we applied some of the most successful convolutional neural networks (ResNet, Inception, VGG, and AlexNet) for 10-way classification of the X-ray images. We showed that substantial classification accuracy is gained by using such networks compared to two simpler ones previously proposed for this purpose. The best accuracy was obtained from ResNet (81.43%), which corresponds to a missed crystal rate of 5.9%. This rate could be lowered to less than 0.1% by using a top-3 classification strategy. Our dataset consisted of 486,000 internally annotated images, which was augmented to more than a million to address class imbalance. We also provide a label-wise analysis of the results, identifying the main sources of error and inaccuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge