Soheil Ghafurian
Group Equivariant Generative Adversarial Networks
May 04, 2020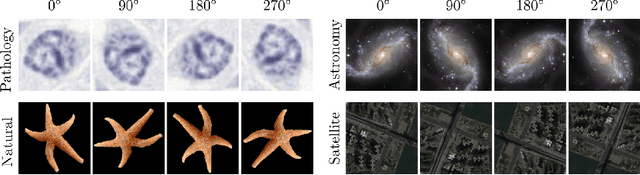
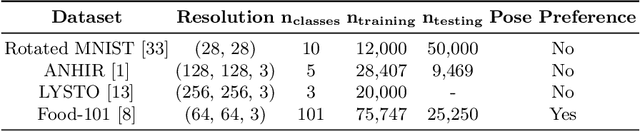
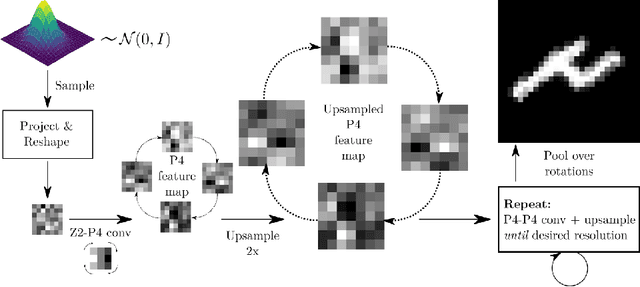
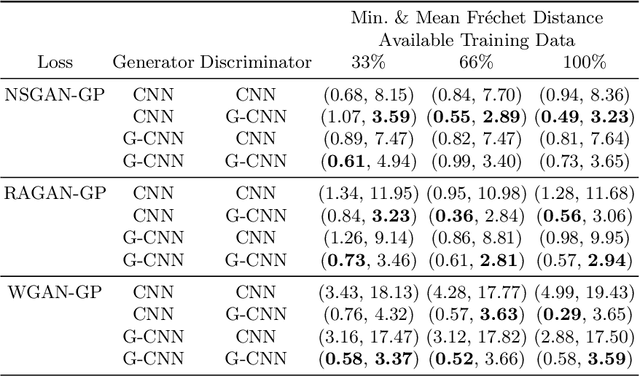
Abstract:Generative adversarial networks are the state of the art for generative modeling in vision, yet are notoriously unstable in practice. This instability is further exacerbated with limited training data. However, in the synthesis of domains such as medical or satellite imaging, it is often overlooked that the image label is invariant to global image symmetries (e.g., rotations and reflections). In this work, we improve gradient feedback between generator and discriminator using an inductive symmetry prior via group-equivariant convolutional networks. We replace convolutional layers with equivalent group-convolutional layers in both generator and discriminator, allowing for better optimization steps and increased expressive power with limited samples. In the process, we extend recent GAN developments to the group-equivariant setting. We demonstrate the utility of our methods by improving both sample fidelity and diversity in the class-conditional synthesis of a diverse set of globally-symmetric imaging modalities.
Restoration of marker occluded hematoxylin and eosin stained whole slide histology images using generative adversarial networks
Oct 14, 2019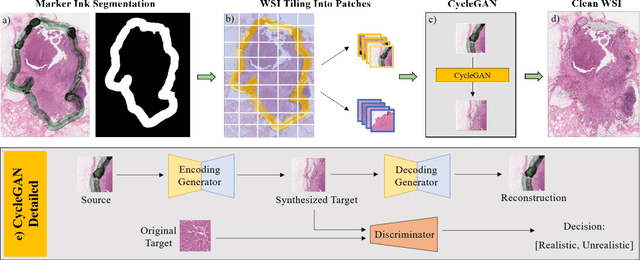
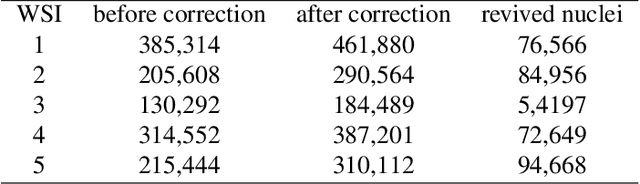
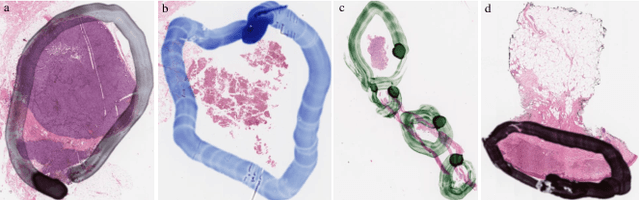
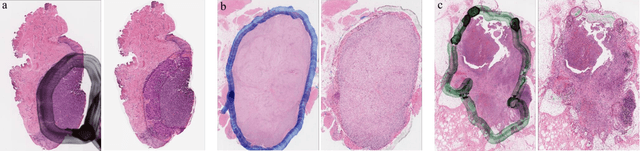
Abstract:It is common for pathologists to annotate specific regions of the tissue, such as tumor, directly on the glass slide with markers. Although this practice was helpful prior to the advent of histology whole slide digitization, it often occludes important details which are increasingly relevant to immuno-oncology due to recent advancements in digital pathology imaging techniques. The current work uses a generative adversarial network with cycle loss to remove these annotations while still maintaining the underlying structure of the tissue by solving an image-to-image translation problem. We train our network on up to 300 whole slide images with marker inks and show that 70% of the corrected image patches are indistinguishable from originally uncontaminated image tissue to a human expert. This portion increases 97% when we replace the human expert with a deep residual network. We demonstrated the fidelity of the method to the original image by calculating the correlation between image gradient magnitudes. We observed a revival of up to 94,000 nuclei per slide in our dataset, the majority of which were located on tissue border.
Classification of Protein Crystallization X-Ray Images Using Major Convolutional Neural Network Architectures
May 11, 2018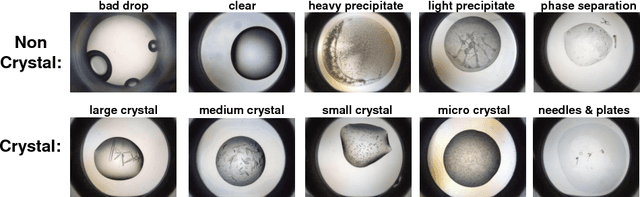
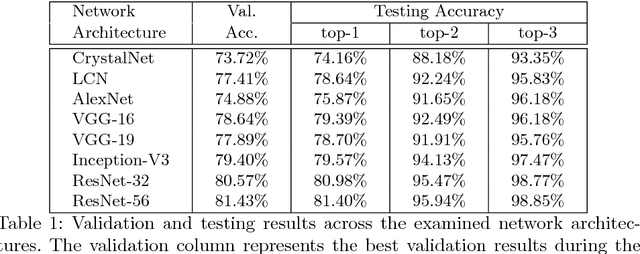
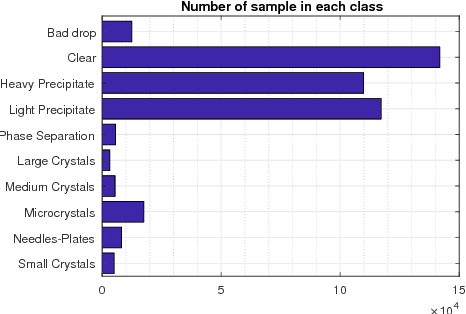
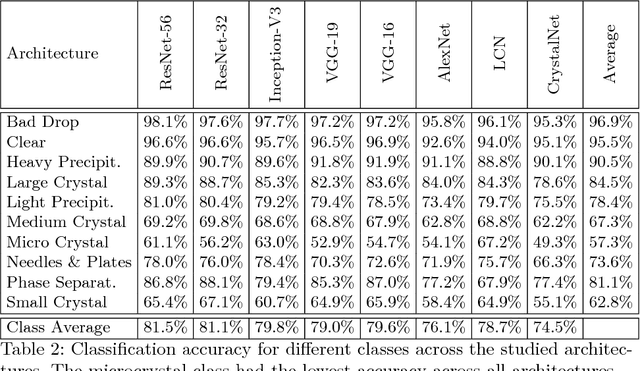
Abstract:The generation of protein crystals is necessary for the study of protein molecular function and structure. This is done empirically by processing large numbers of crystallization trials and inspecting them regularly in search of those with forming crystals. To avoid missing the hard-gained crystals, this visual inspection of the trial X-ray images is done manually as opposed to the existing less accurate machine learning methods. To achieve higher accuracy for automation, we applied some of the most successful convolutional neural networks (ResNet, Inception, VGG, and AlexNet) for 10-way classification of the X-ray images. We showed that substantial classification accuracy is gained by using such networks compared to two simpler ones previously proposed for this purpose. The best accuracy was obtained from ResNet (81.43%), which corresponds to a missed crystal rate of 5.9%. This rate could be lowered to less than 0.1% by using a top-3 classification strategy. Our dataset consisted of 486,000 internally annotated images, which was augmented to more than a million to address class imbalance. We also provide a label-wise analysis of the results, identifying the main sources of error and inaccuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge