Belma Dogdas
A multi-level convolutional LSTM model for the segmentation of left ventricle myocardium in infarcted porcine cine MR images
Nov 14, 2018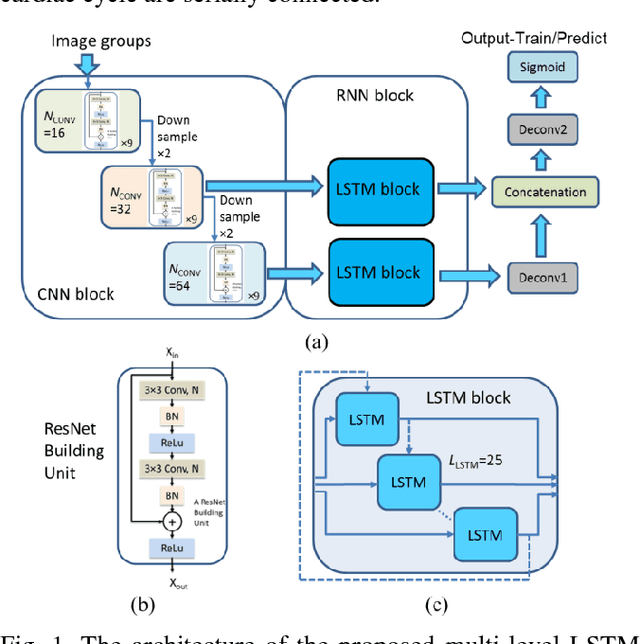
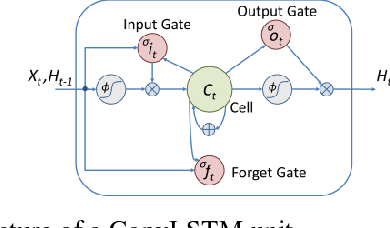
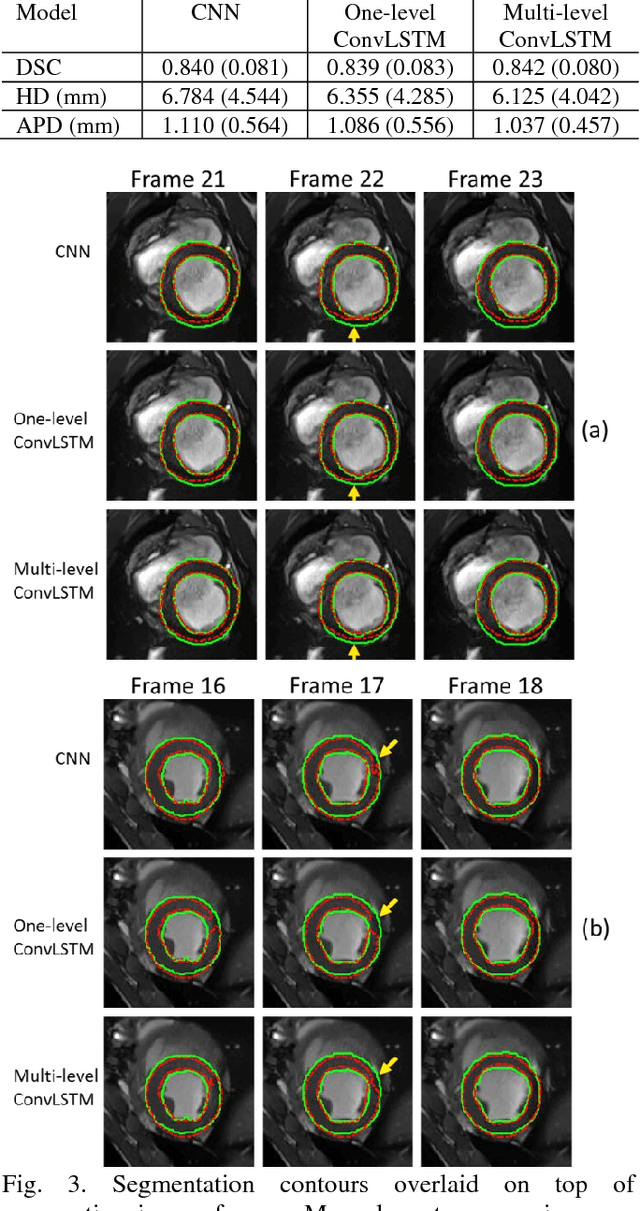
Abstract:Automatic segmentation of left ventricle (LV) myocardium in cardiac short-axis cine MR images acquired on subjects with myocardial infarction is a challenging task, mainly because of the various types of image inhomogeneity caused by the infarctions. Among the approaches proposed to automate the LV myocardium segmentation task, methods based upon deep convolutional neural networks (CNN) have demonstrated their exceptional accuracy and robustness in recent years. However, most of the CNN-based approaches treat the frames in a cardiac cycle independently, which fails to capture the valuable dynamics of heart motion. Herein, an approach based on recurrent neural network (RNN), specifically a multi-level convolutional long short-term memory (ConvLSTM) model, is proposed to take the motion of the heart into consideration. Based on a ResNet-56 CNN, LV-related image features in consecutive frames of a cardiac cycle are extracted at both the low- and high-resolution levels, which are processed by the corresponding multi-level ConvLSTM models to generate the myocardium segmentations. A leave-one-out experiment was carried out on a set of 3,600 cardiac cine MR slices collected in-house for 8 porcine subjects with surgically induced myocardial infarction. Compared with a solely CNN-based approach, the proposed approach demonstrated its superior robustness against image inhomogeneity by incorporating information from adjacent frames. It also outperformed a one-level ConvLSTM approach thanks to its capabilities to take advantage of image features at multiple resolution levels.
A Progressively-trained Scale-invariant and Boundary-aware Deep Neural Network for the Automatic 3D Segmentation of Lung Lesions
Nov 11, 2018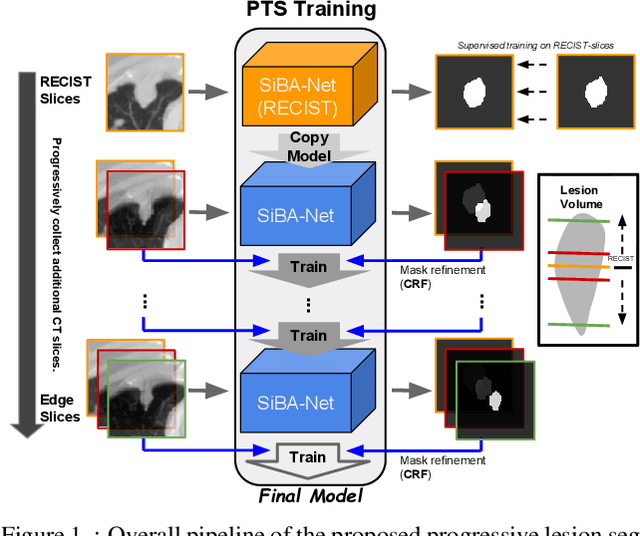
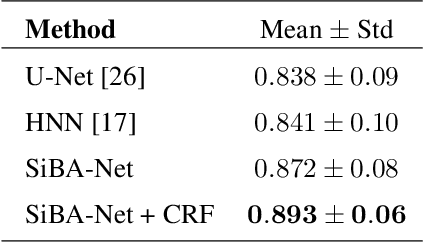
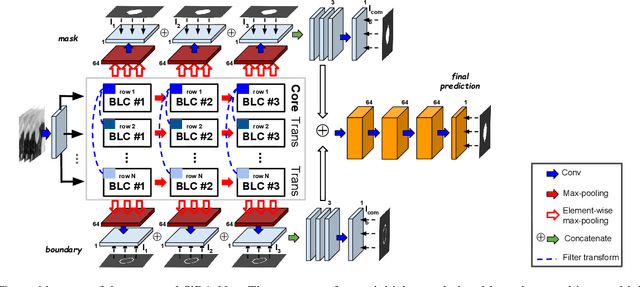
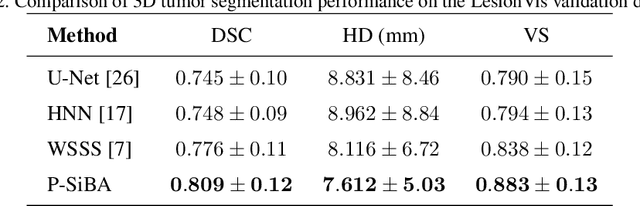
Abstract:Volumetric segmentation of lesions on CT scans is important for many types of analysis, including lesion growth kinetic modeling in clinical trials and machine learning of radiomic features. Manual segmentation is laborious, and impractical for large-scale use. For routine clinical use, and in clinical trials that apply the Response Evaluation Criteria In Solid Tumors (RECIST), clinicians typically outline the boundaries of a lesion on a single slice to extract diameter measurements. In this work, we have collected a large-scale database, named LesionVis, with pixel-wise manual 2D lesion delineations on the RECIST-slices. To extend the 2D segmentations to 3D, we propose a volumetric progressive lesion segmentation (PLS) algorithm to automatically segment the 3D lesion volume from 2D delineations using a scale-invariant and boundary-aware deep convolutional network (SIBA-Net). The SIBA-Net copes with the size transition of a lesion when the PLS progresses from the RECIST-slice to the edge-slices, as well as when performing longitudinal assessment of lesions whose size change over multiple time points. The proposed PLS-SiBA-Net (P-SiBA) approach is assessed on the lung lesion cases from LesionVis. Our experimental results demonstrate that the P-SiBA approach achieves mean Dice similarity coefficients (DSC) of 0.81, which significantly improves 3D segmentation accuracy compared with the approaches proposed previously (highest mean DSC at 0.78 on LesionVis). In summary, by leveraging the limited 2D delineations on the RECIST-slices, P-SiBA is an effective semi-supervised approach to produce accurate lesion segmentations in 3D.
Classification of Protein Crystallization X-Ray Images Using Major Convolutional Neural Network Architectures
May 11, 2018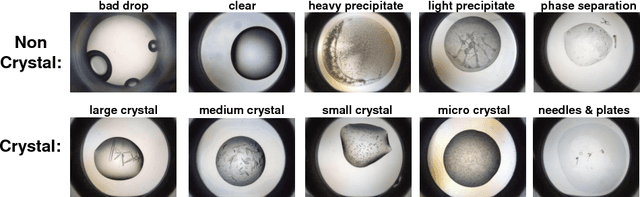
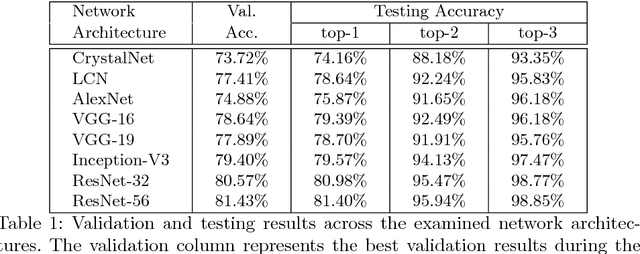
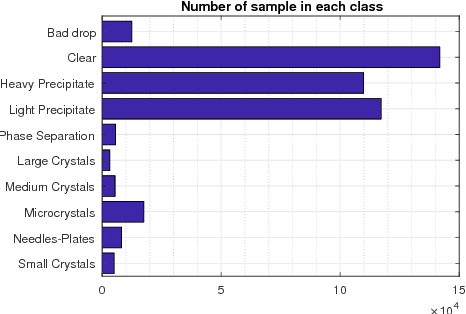
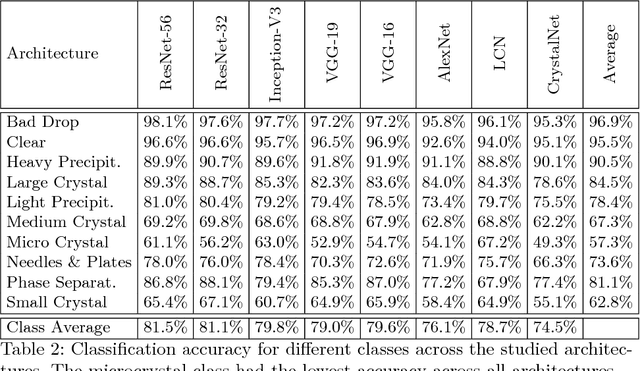
Abstract:The generation of protein crystals is necessary for the study of protein molecular function and structure. This is done empirically by processing large numbers of crystallization trials and inspecting them regularly in search of those with forming crystals. To avoid missing the hard-gained crystals, this visual inspection of the trial X-ray images is done manually as opposed to the existing less accurate machine learning methods. To achieve higher accuracy for automation, we applied some of the most successful convolutional neural networks (ResNet, Inception, VGG, and AlexNet) for 10-way classification of the X-ray images. We showed that substantial classification accuracy is gained by using such networks compared to two simpler ones previously proposed for this purpose. The best accuracy was obtained from ResNet (81.43%), which corresponds to a missed crystal rate of 5.9%. This rate could be lowered to less than 0.1% by using a top-3 classification strategy. Our dataset consisted of 486,000 internally annotated images, which was augmented to more than a million to address class imbalance. We also provide a label-wise analysis of the results, identifying the main sources of error and inaccuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge