Hillary Ngai
Mah Parsa
Audio-AdapterFusion: A Task-ID-free Approach for Efficient and Non-Destructive Multi-task Speech Recognition
Oct 17, 2023Abstract:Adapters are an efficient, composable alternative to full fine-tuning of pre-trained models and help scale the deployment of large ASR models to many tasks. In practice, a task ID is commonly prepended to the input during inference to route to single-task adapters for the specified task. However, one major limitation of this approach is that the task ID may not be known during inference, rendering it unsuitable for most multi-task settings. To address this, we propose three novel task-ID-free methods to combine single-task adapters in multi-task ASR and investigate two learning algorithms for training. We evaluate our methods on 10 test sets from 4 diverse ASR tasks and show that our methods are non-destructive and parameter-efficient. While only updating 17% of the model parameters, our methods can achieve an 8% mean WER improvement relative to full fine-tuning and are on-par with task-ID adapter routing.
Doctor XAvIer: Explainable Diagnosis on Physician-Patient Dialogues and XAI Evaluation
Apr 25, 2022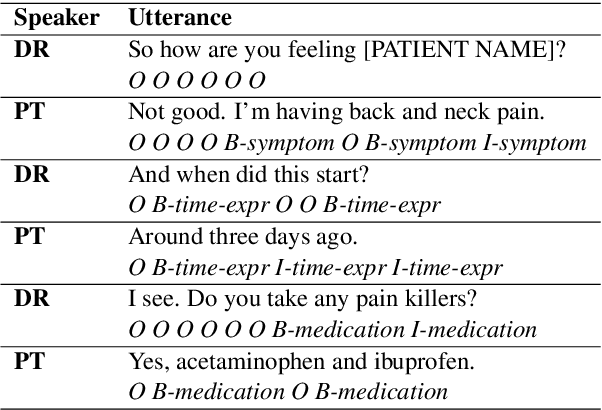

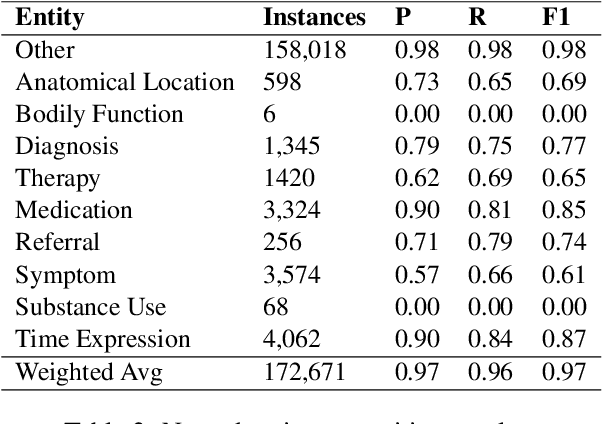
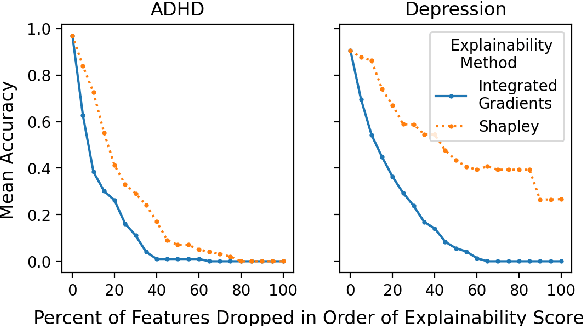
Abstract:We introduce Doctor XAvIer, a BERT-based diagnostic system that extracts relevant clinical data from transcribed patient-doctor dialogues and explains predictions using feature attribution methods. We present a novel performance plot and evaluation metric for feature attribution methods: Feature Attribution Dropping (FAD) curve and its Normalized Area Under the Curve (N-AUC). FAD curve analysis shows that integrated gradients outperforms Shapley values in explaining diagnosis classification. Doctor XAvIer outperforms the baseline with 0.97 F1-score in named entity recognition and symptom pertinence classification and 0.91 F1-score in diagnosis classification.
Transformer-Based Models for Question Answering on COVID19
Jan 16, 2021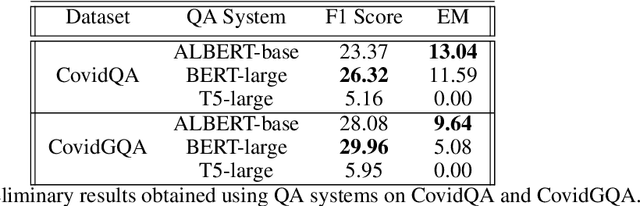
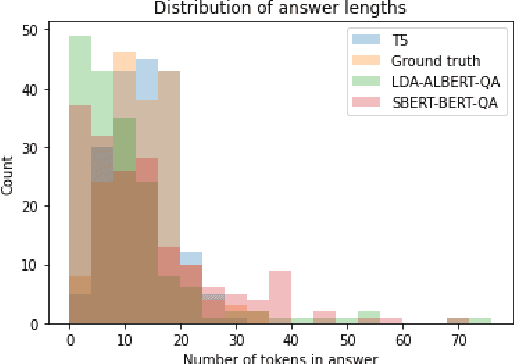
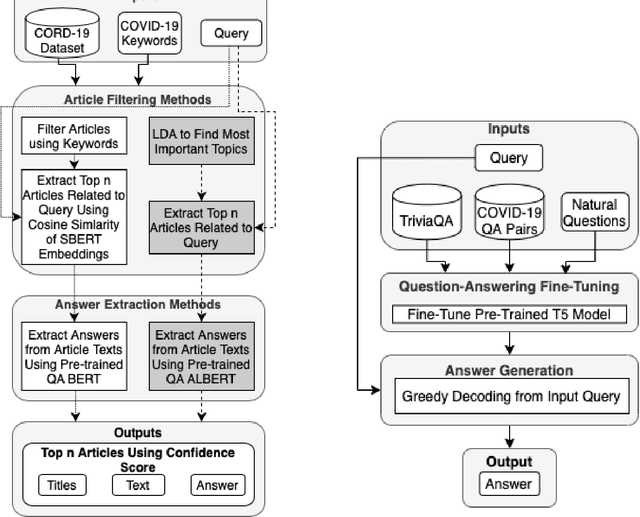
Abstract:In response to the Kaggle's COVID-19 Open Research Dataset (CORD-19) challenge, we have proposed three transformer-based question-answering systems using BERT, ALBERT, and T5 models. Since the CORD-19 dataset is unlabeled, we have evaluated the question-answering models' performance on two labeled questions answers datasets \textemdash CovidQA and CovidGQA. The BERT-based QA system achieved the highest F1 score (26.32), while the ALBERT-based QA system achieved the highest Exact Match (13.04). However, numerous challenges are associated with developing high-performance question-answering systems for the ongoing COVID-19 pandemic and future pandemics. At the end of this paper, we discuss these challenges and suggest potential solutions to address them.
An Experimental Evaluation of Transformer-based Language Models in the Biomedical Domain
Dec 31, 2020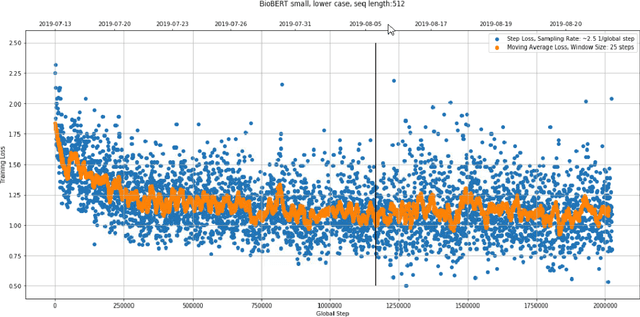
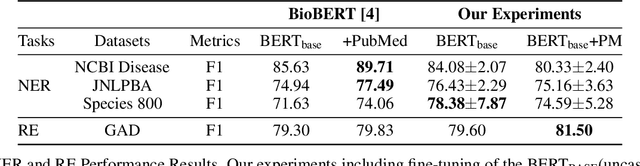
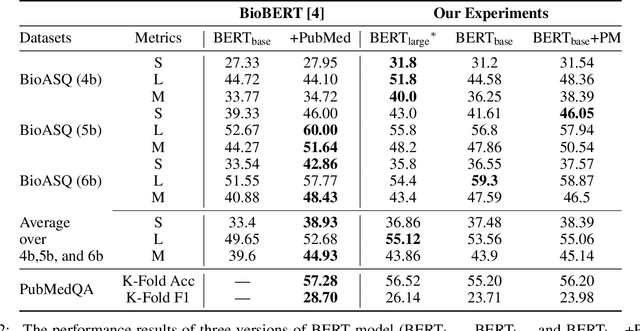
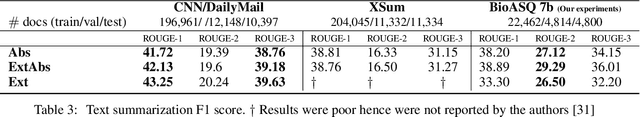
Abstract:With the growing amount of text in health data, there have been rapid advances in large pre-trained models that can be applied to a wide variety of biomedical tasks with minimal task-specific modifications. Emphasizing the cost of these models, which renders technical replication challenging, this paper summarizes experiments conducted in replicating BioBERT and further pre-training and careful fine-tuning in the biomedical domain. We also investigate the effectiveness of domain-specific and domain-agnostic pre-trained models across downstream biomedical NLP tasks. Our finding confirms that pre-trained models can be impactful in some downstream NLP tasks (QA and NER) in the biomedical domain; however, this improvement may not justify the high cost of domain-specific pre-training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge