Gyumin Lee
Repurformer: Transformers for Repurposing-Aware Molecule Generation
Jul 16, 2024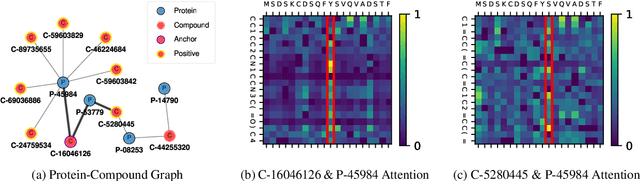
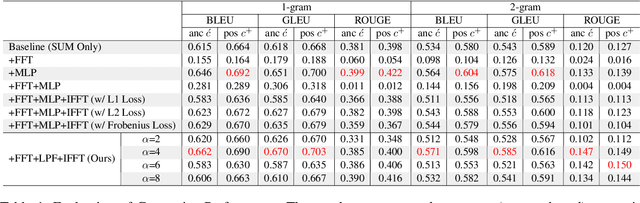
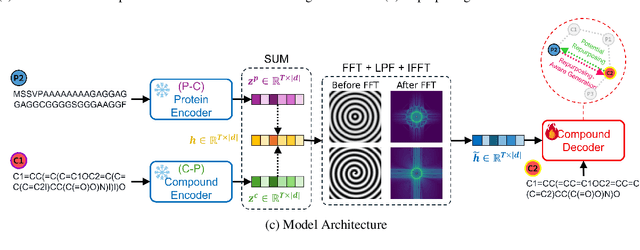
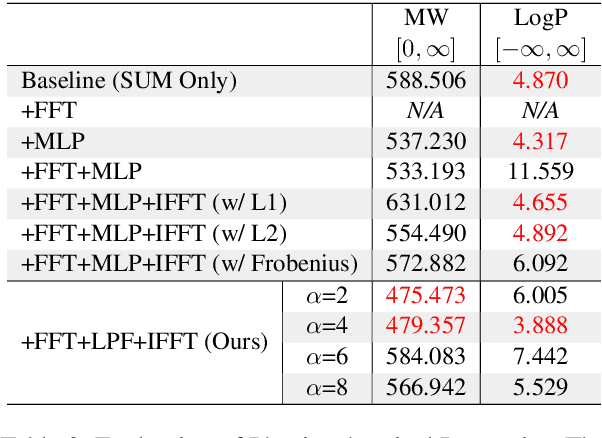
Abstract:Generating as diverse molecules as possible with desired properties is crucial for drug discovery research, which invokes many approaches based on deep generative models today. Despite recent advancements in these models, particularly in variational autoencoders (VAEs), generative adversarial networks (GANs), Transformers, and diffusion models, a significant challenge known as \textit{the sample bias problem} remains. This problem occurs when generated molecules targeting the same protein tend to be structurally similar, reducing the diversity of generation. To address this, we propose leveraging multi-hop relationships among proteins and compounds. Our model, Repurformer, integrates bi-directional pretraining with Fast Fourier Transform (FFT) and low-pass filtering (LPF) to capture complex interactions and generate diverse molecules. A series of experiments on BindingDB dataset confirm that Repurformer successfully creates substitutes for anchor compounds that resemble positive compounds, increasing diversity between the anchor and generated compounds.
Time Series Forecasting with Hypernetworks Generating Parameters in Advance
Nov 22, 2022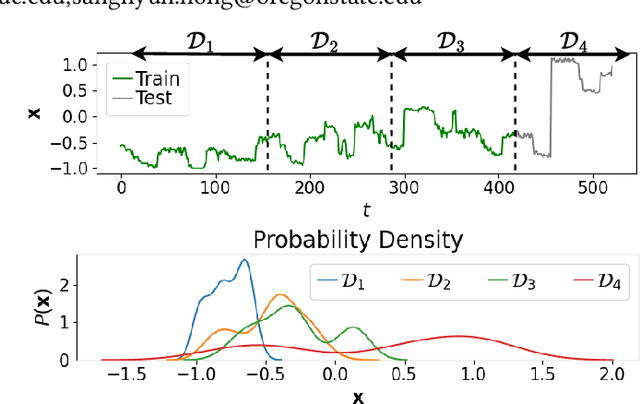

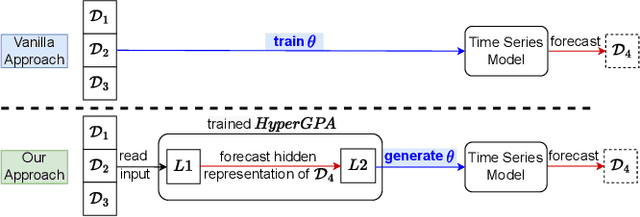
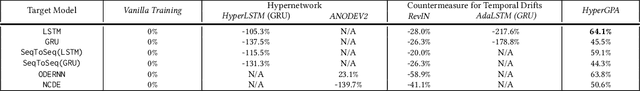
Abstract:Forecasting future outcomes from recent time series data is not easy, especially when the future data are different from the past (i.e. time series are under temporal drifts). Existing approaches show limited performances under data drifts, and we identify the main reason: It takes time for a model to collect sufficient training data and adjust its parameters for complicated temporal patterns whenever the underlying dynamics change. To address this issue, we study a new approach; instead of adjusting model parameters (by continuously re-training a model on new data), we build a hypernetwork that generates other target models' parameters expected to perform well on the future data. Therefore, we can adjust the model parameters beforehand (if the hypernetwork is correct). We conduct extensive experiments with 6 target models, 6 baselines, and 4 datasets, and show that our HyperGPA outperforms other baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge