Gianmaria Silvello
A Domain-Specific Curated Benchmark for Entity and Document-Level Relation Extraction
Feb 04, 2026Abstract:Information Extraction (IE), encompassing Named Entity Recognition (NER), Named Entity Linking (NEL), and Relation Extraction (RE), is critical for transforming the rapidly growing volume of scientific publications into structured, actionable knowledge. This need is especially evident in fast-evolving biomedical fields such as the gut-brain axis, where research investigates complex interactions between the gut microbiota and brain-related disorders. Existing biomedical IE benchmarks, however, are often narrow in scope and rely heavily on distantly supervised or automatically generated annotations, limiting their utility for advancing robust IE methods. We introduce GutBrainIE, a benchmark based on more than 1,600 PubMed abstracts, manually annotated by biomedical and terminological experts with fine-grained entities, concept-level links, and relations. While grounded in the gut-brain axis, the benchmark's rich schema, multiple tasks, and combination of highly curated and weakly supervised data make it broadly applicable to the development and evaluation of biomedical IE systems across domains.
DOREMI: Optimizing Long Tail Predictions in Document-Level Relation Extraction
Jan 16, 2026Abstract:Document-Level Relation Extraction (DocRE) presents significant challenges due to its reliance on cross-sentence context and the long-tail distribution of relation types, where many relations have scarce training examples. In this work, we introduce DOcument-level Relation Extraction optiMizing the long taIl (DOREMI), an iterative framework that enhances underrepresented relations through minimal yet targeted manual annotations. Unlike previous approaches that rely on large-scale noisy data or heuristic denoising, DOREMI actively selects the most informative examples to improve training efficiency and robustness. DOREMI can be applied to any existing DocRE model and is effective at mitigating long-tail biases, offering a scalable solution to improve generalization on rare relations.
From Single to Multi-Agent Reasoning: Advancing GeneGPT for Genomics QA
Jan 15, 2026Abstract:Comprehending genomic information is essential for biomedical research, yet extracting data from complex distributed databases remains challenging. Large language models (LLMs) offer potential for genomic Question Answering (QA) but face limitations due to restricted access to domain-specific databases. GeneGPT is the current state-of-the-art system that enhances LLMs by utilizing specialized API calls, though it is constrained by rigid API dependencies and limited adaptability. We replicate GeneGPT and propose GenomAgent, a multi-agent framework that efficiently coordinates specialized agents for complex genomics queries. Evaluated on nine tasks from the GeneTuring benchmark, GenomAgent outperforms GeneGPT by 12% on average, and its flexible architecture extends beyond genomics to various scientific domains needing expert knowledge extraction.
Efficient and Reliable Estimation of Named Entity Linking Quality: A Case Study on GutBrainIE
Jan 10, 2026Abstract:Named Entity Linking (NEL) is a core component of biomedical Information Extraction (IE) pipelines, yet assessing its quality at scale is challenging due to the high cost of expert annotations and the large size of corpora. In this paper, we present a sampling-based framework to estimate the NEL accuracy of large-scale IE corpora under statistical guarantees and constrained annotation budgets. We frame NEL accuracy estimation as a constrained optimization problem, where the objective is to minimize expected annotation cost subject to a target Margin of Error (MoE) for the corpus-level accuracy estimate. Building on recent works on knowledge graph accuracy estimation, we adapt Stratified Two-Stage Cluster Sampling (STWCS) to the NEL setting, defining label-based strata and global surface-form clusters in a way that is independent of NEL annotations. Applied to 11,184 NEL annotations in GutBrainIE -- a new biomedical corpus openly released in fall 2025 -- our framework reaches a MoE $\leq 0.05$ by manually annotating only 2,749 triples (24.6%), leading to an overall accuracy estimate of $0.915 \pm 0.0473$. A time-based cost model and simulations against a Simple Random Sampling (SRS) baseline show that our design reduces expert annotation time by about 29% at fixed sample size. The framework is generic and can be applied to other NEL benchmarks and IE pipelines that require scalable and statistically robust accuracy assessment.
Testing software for non-discrimination: an updated and extended audit in the Italian car insurance domain
Feb 10, 2025Abstract:Context. As software systems become more integrated into society's infrastructure, the responsibility of software professionals to ensure compliance with various non-functional requirements increases. These requirements include security, safety, privacy, and, increasingly, non-discrimination. Motivation. Fairness in pricing algorithms grants equitable access to basic services without discriminating on the basis of protected attributes. Method. We replicate a previous empirical study that used black box testing to audit pricing algorithms used by Italian car insurance companies, accessible through a popular online system. With respect to the previous study, we enlarged the number of tests and the number of demographic variables under analysis. Results. Our work confirms and extends previous findings, highlighting the problematic permanence of discrimination across time: demographic variables significantly impact pricing to this day, with birthplace remaining the main discriminatory factor against individuals not born in Italian cities. We also found that driver profiles can determine the number of quotes available to the user, denying equal opportunities to all. Conclusion. The study underscores the importance of testing for non-discrimination in software systems that affect people's everyday lives. Performing algorithmic audits over time makes it possible to evaluate the evolution of such algorithms. It also demonstrates the role that empirical software engineering can play in making software systems more accountable.
Automatic Labels are as Effective as Manual Labels in Biomedical Images Classification with Deep Learning
Jun 20, 2024Abstract:The increasing availability of biomedical data is helping to design more robust deep learning (DL) algorithms to analyze biomedical samples. Currently, one of the main limitations to train DL algorithms to perform a specific task is the need for medical experts to label data. Automatic methods to label data exist, however automatic labels can be noisy and it is not completely clear when automatic labels can be adopted to train DL models. This paper aims to investigate under which circumstances automatic labels can be adopted to train a DL model on the classification of Whole Slide Images (WSI). The analysis involves multiple architectures, such as Convolutional Neural Networks (CNN) and Vision Transformer (ViT), and over 10000 WSIs, collected from three use cases: celiac disease, lung cancer and colon cancer, which one including respectively binary, multiclass and multilabel data. The results allow identifying 10% as the percentage of noisy labels that lead to train competitive models for the classification of WSIs. Therefore, an algorithm generating automatic labels needs to fit this criterion to be adopted. The application of the Semantic Knowledge Extractor Tool (SKET) algorithm to generate automatic labels leads to performance comparable to the one obtained with manual labels, since it generates a percentage of noisy labels between 2-5%. Automatic labels are as effective as manual ones, reaching solid performance comparable to the one obtained training models with manual labels.
Learning to Rank from Relevance Judgments Distributions
Feb 13, 2022



Abstract:Learning to Rank (LETOR) algorithms are usually trained on annotated corpora where a single relevance label is assigned to each available document-topic pair. Within the Cranfield framework, relevance labels result from merging either multiple expertly curated or crowdsourced human assessments. In this paper, we explore how to train LETOR models with relevance judgments distributions (either real or synthetically generated) assigned to document-topic pairs instead of single-valued relevance labels. We propose five new probabilistic loss functions to deal with the higher expressive power provided by relevance judgments distributions and show how they can be applied both to neural and GBM architectures. Moreover, we show how training a LETOR model on a sampled version of the relevance judgments from certain probability distributions can improve its performance when relying either on traditional or probabilistic loss functions. Finally, we validate our hypothesis on real-world crowdsourced relevance judgments distributions. Overall, we observe that relying on relevance judgments distributions to train different LETOR models can boost their performance and even outperform strong baselines such as LambdaMART on several test collections.
Neural Feature Selection for Learning to Rank
Feb 22, 2021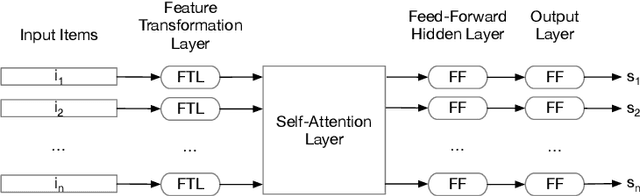
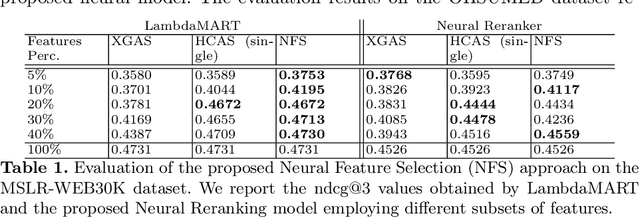

Abstract:LEarning TO Rank (LETOR) is a research area in the field of Information Retrieval (IR) where machine learning models are employed to rank a set of items. In the past few years, neural LETOR approaches have become a competitive alternative to traditional ones like LambdaMART. However, neural architectures performance grew proportionally to their complexity and size. This can be an obstacle for their adoption in large-scale search systems where a model size impacts latency and update time. For this reason, we propose an architecture-agnostic approach based on a neural LETOR model to reduce the size of its input by up to 60% without affecting the system performance. This approach also allows to reduce a LETOR model complexity and, therefore, its training and inference time up to 50%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge