Georg Rose
Institute for Medical Engineering and Research Campus STIMULATE, University of Magdeburg, Magdeburg, Germany
Liver Segmentation in Time-resolved C-arm CT Volumes Reconstructed from Dynamic Perfusion Scans using Time Separation Technique
Feb 09, 2023



Abstract:Perfusion imaging is a valuable tool for diagnosing and treatment planning for liver tumours. The time separation technique (TST) has been successfully used for modelling C-arm cone-beam computed tomography (CBCT) perfusion data. The reconstruction can be accompanied by the segmentation of the liver - for better visualisation and for generating comprehensive perfusion maps. Recently introduced Turbolift learning has been seen to perform well while working with TST reconstructions, but has not been explored for the time-resolved volumes (TRV) estimated out of TST reconstructions. The segmentation of the TRVs can be useful for tracking the movement of the liver over time. This research explores this possibility by training the multi-scale attention UNet of Turbolift learning at its third stage on the TRVs and shows the robustness of Turbolift learning since it can even work efficiently with the TRVs, resulting in a Dice score of 0.864$\pm$0.004.
Liver Segmentation using Turbolift Learning for CT and Cone-beam C-arm Perfusion Imaging
Jul 20, 2022
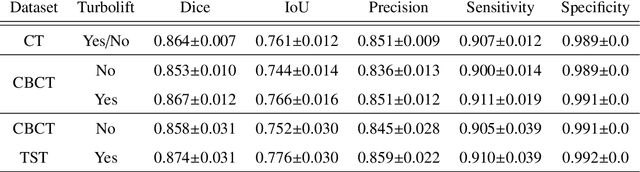
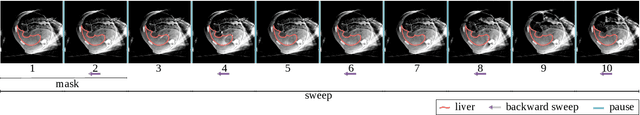
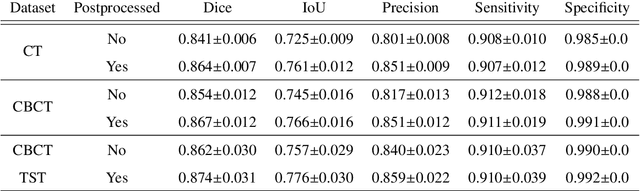
Abstract:Model-based reconstruction employing the time separation technique (TST) was found to improve dynamic perfusion imaging of the liver using C-arm cone-beam computed tomography (CBCT). To apply TST using prior knowledge extracted from CT perfusion data, the liver should be accurately segmented from the CT scans. Reconstructions of primary and model-based CBCT data need to be segmented for proper visualisation and interpretation of perfusion maps. This research proposes Turbolift learning, which trains a modified version of the multi-scale Attention UNet on different liver segmentation tasks serially, following the order of the trainings CT, CBCT, CBCT TST - making the previous trainings act as pre-training stages for the subsequent ones - addressing the problem of limited number of datasets for training. For the final task of liver segmentation from CBCT TST, the proposed method achieved an overall Dice scores of 0.874$\pm$0.031 and 0.905$\pm$0.007 in 6-fold and 4-fold cross-validation experiments, respectively - securing statistically significant improvements over the model, which was trained only for that task. Experiments revealed that Turbolift not only improves the overall performance of the model but also makes it robust against artefacts originating from the embolisation materials and truncation artefacts. Additionally, in-depth analyses confirmed the order of the segmentation tasks. This paper shows the potential of segmenting the liver from CT, CBCT, and CBCT TST, learning from the available limited training data, which can possibly be used in the future for the visualisation and evaluation of the perfusion maps for the treatment evaluation of liver diseases.
Primal-Dual UNet for Sparse View Cone Beam Computed Tomography Volume Reconstruction
May 11, 2022
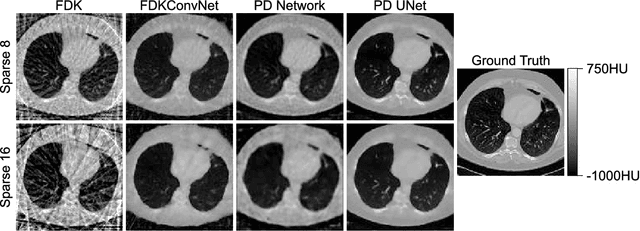
Abstract:In this paper, the Primal-Dual UNet for sparse view CT reconstruction is modified to be applicable to cone beam projections and perform reconstructions of entire volumes instead of slices. Experiments show that the PSNR of the proposed method is increased by 10dB compared to the direct FDK reconstruction and almost 3dB compared to the modified original Primal-Dual Network when using only 23 projections. The presented network is not optimized wrt. memory consumption or hyperparameters but merely serves as a proof of concept and is limited to low resolution projections and volumes.
Dual Branch Prior-SegNet: CNN for Interventional CBCT using Planning Scan and Auxiliary Segmentation Loss
May 11, 2022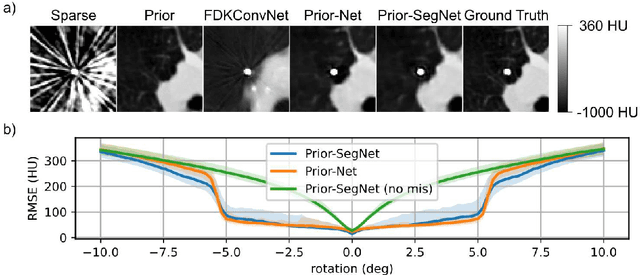
Abstract:This paper proposes an extension to the Dual Branch Prior-Net for sparse view interventional CBCT reconstruction incorporating a high quality planning scan. An additional head learns to segment interventional instruments and thus guides the reconstruction task. The prior scans are misaligned by up to +-5deg in-plane during training. Experiments show that the proposed model, Dual Branch Prior-SegNet, significantly outperforms any other evaluated model by >2.8dB PSNR. It also stays robust wrt. rotations of up to +-5.5deg.
DDoS-UNet: Incorporating temporal information using Dynamic Dual-channel UNet for enhancing super-resolution of dynamic MRI
Feb 10, 2022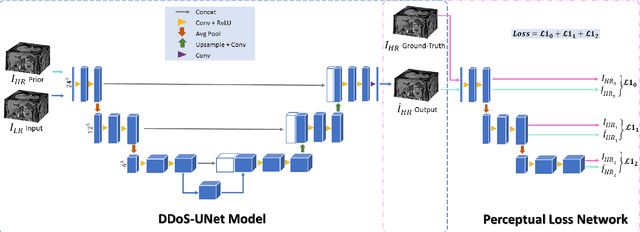
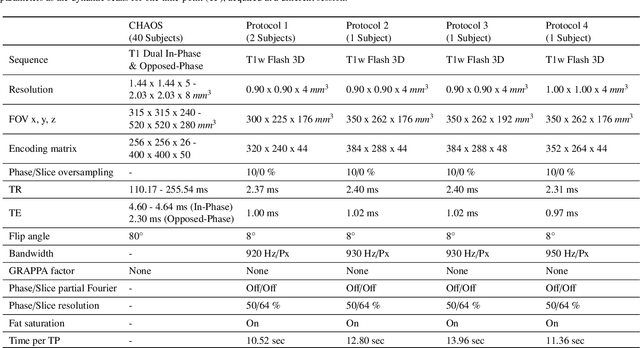
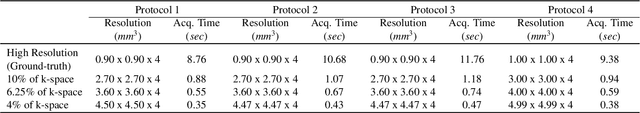
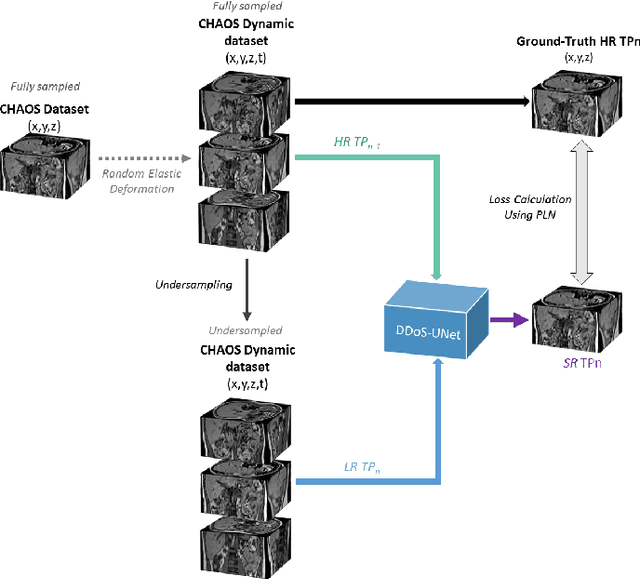
Abstract:Magnetic resonance imaging (MRI) provides high spatial resolution and excellent soft-tissue contrast without using harmful ionising radiation. Dynamic MRI is an essential tool for interventions to visualise movements or changes of the target organ. However, such MRI acquisition with high temporal resolution suffers from limited spatial resolution - also known as the spatio-temporal trade-off of dynamic MRI. Several approaches, including deep learning based super-resolution approaches, have been proposed to mitigate this trade-off. Nevertheless, such an approach typically aims to super-resolve each time-point separately, treating them as individual volumes. This research addresses the problem by creating a deep learning model which attempts to learn both spatial and temporal relationships. A modified 3D UNet model, DDoS-UNet, is proposed - which takes the low-resolution volume of the current time-point along with a prior image volume. Initially, the network is supplied with a static high-resolution planning scan as the prior image along with the low-resolution input to super-resolve the first time-point. Then it continues step-wise by using the super-resolved time-points as the prior image while super-resolving the subsequent time-points. The model performance was tested with 3D dynamic data that was undersampled to different in-plane levels. The proposed network achieved an average SSIM value of 0.951$\pm$0.017 while reconstructing the lowest resolution data (i.e. only 4\% of the k-space acquired) - which could result in a theoretical acceleration factor of 25. The proposed approach can be used to reduce the required scan-time while achieving high spatial resolution.
Sinogram upsampling using Primal-Dual UNet for undersampled CT and radial MRI reconstruction
Dec 26, 2021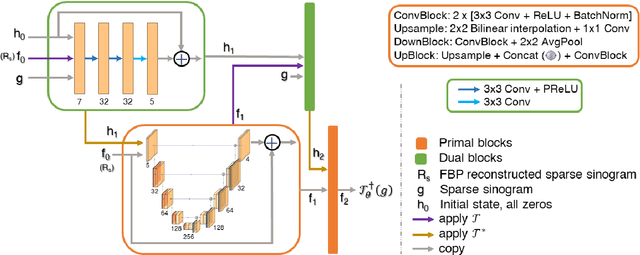
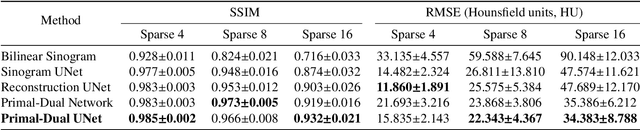
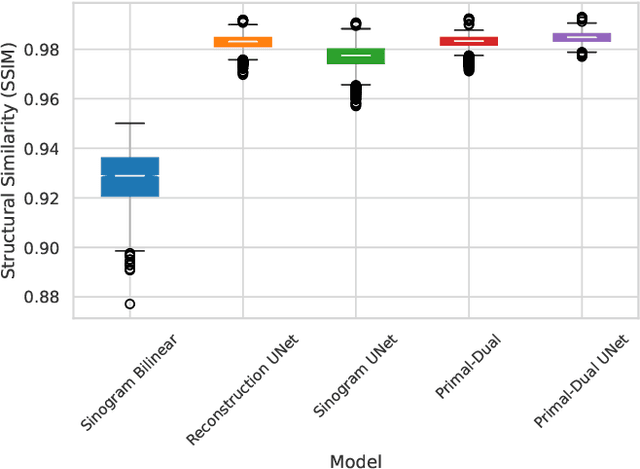
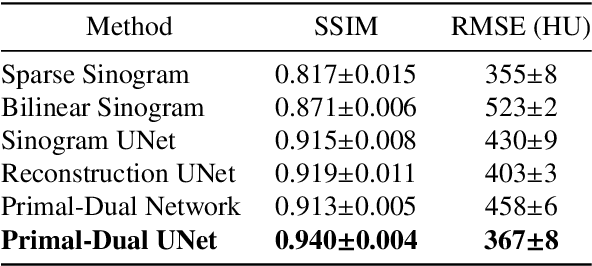
Abstract:CT and MRI are two widely used clinical imaging modalities for non-invasive diagnosis. However, both of these modalities come with certain problems. CT uses harmful ionising radiation, and MRI suffers from slow acquisition speed. Both problems can be tackled by undersampling, such as sparse sampling. However, such undersampled data leads to lower resolution and introduces artefacts. Several techniques, including deep learning based methods, have been proposed to reconstruct such data. However, the undersampled reconstruction problem for these two modalities was always considered as two different problems and tackled separately by different research works. This paper proposes a unified solution for both sparse CT and undersampled radial MRI reconstruction, achieved by applying Fourier transform-based pre-processing on the radial MRI and then reconstructing both modalities using sinogram upsampling combined with filtered back-projection. The Primal-Dual network is a deep learning based method for reconstructing sparsely-sampled CT data. This paper introduces Primal-Dual UNet, which improves the Primal-Dual network in terms of accuracy and reconstruction speed. The proposed method resulted in an average SSIM of 0.932 while performing sparse CT reconstruction for fan-beam geometry with a sparsity level of 16, achieving a statistically significant improvement over the previous model, which resulted in 0.919. Furthermore, the proposed model resulted in 0.903 and 0.957 average SSIM while reconstructing undersampled brain and abdominal MRI data with an acceleration factor of 16 - statistically significant improvements over the original model, which resulted in 0.867 and 0.949. Finally, this paper shows that the proposed network not only improves the overall image quality, but also improves the image quality for the regions-of-interest; as well as generalises better in presence of a needle.
Noise and dose reduction in CT brain perfusion acquisition by projecting time attenuation curves onto lower dimensional spaces
Nov 02, 2021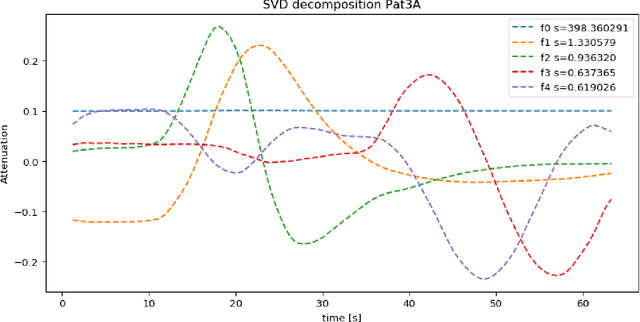
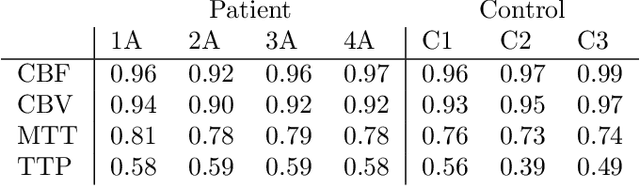
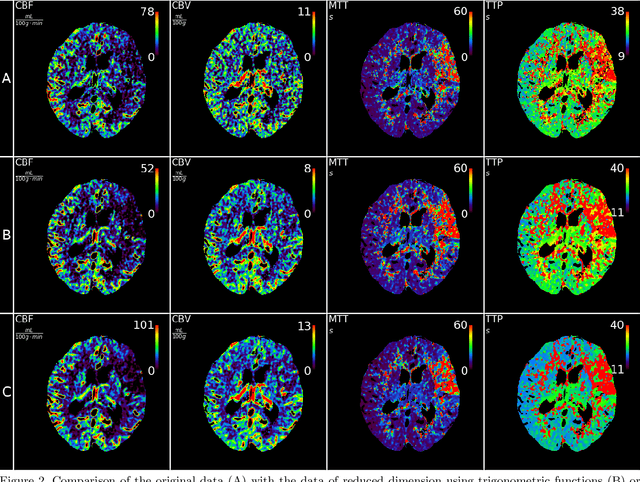

Abstract:CT perfusion imaging (CTP) plays an important role in decision making for the treatment of acute ischemic stroke with large vessel occlusion. Since the CT perfusion scan time is approximately one minute, the patient is exposed to a non-negligible dose of ionizing radiation. However, further dose reduction increases the level of noise in the data and the resulting perfusion maps. We present a method for reducing noise in perfusion data based on dimension reduction of time attenuation curves. For dimension reduction, we use either the fit of the first five terms of the trigonometric polynomial or the first five terms of the SVD decomposition of the time attenuation profiles. CTP data from four patients with large vessel occlusion and three control subjects were studied. To compare the noise level in the perfusion maps, we use the wavelet estimation of the noise standard deviation implemented in the scikit-image package. We show that both methods significantly reduce noise in the data while preserving important information about the perfusion deficits. These methods can be used to further reduce the dose in CT perfusion protocols or in perfusion studies using C-arm CT, which are burdened by high noise levels.
Application of Time Separation Technique to Enhance C-arm CT Dynamic Liver Perfusion Imaging
Oct 27, 2021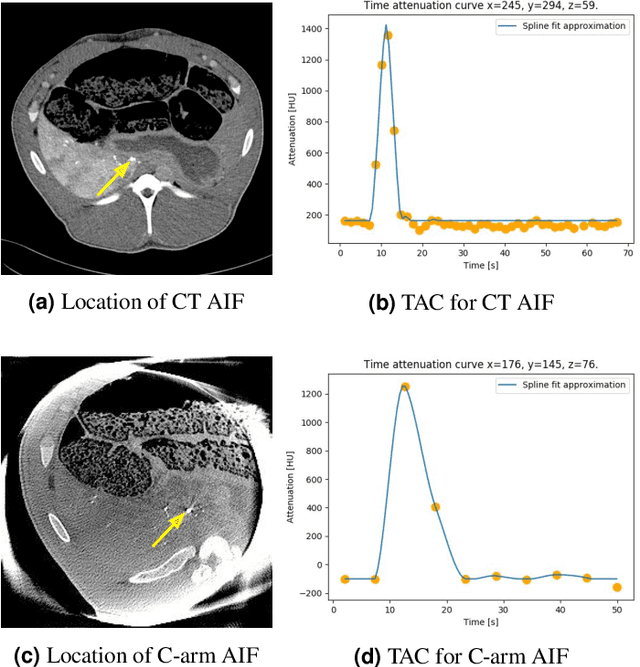
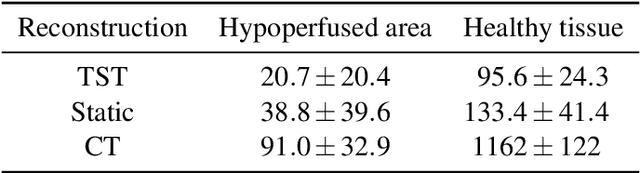
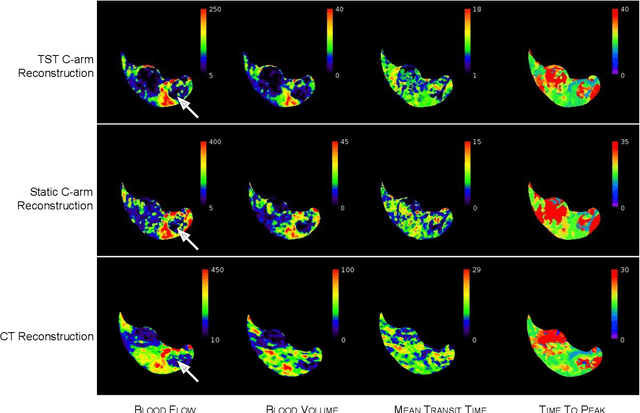
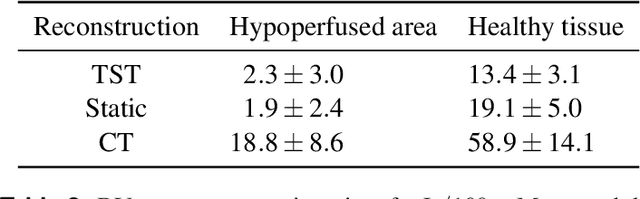
Abstract:Perfusion imaging is an interesting new modality for evaluation and assessment of the liver cancer treatment. C-Arm CT provides a possibility to perform perfusion imaging scans intra-operatively for even faster evaluation. The slow speed of the C-Arm CT rotation and the presence of the noise, however, have an impact on the reconstruction and therefore model based approaches have to be applied. In this work we apply the Time separation technique (TST), to denoise data, speed up reconstruction and improve resulting perfusion images. We show on animal experiment data that Dynamic C-Arm CT Liver Perfusion Imaging together with the processing of the data based on the TST provides comparable results to standard CT liver perfusion imaging.
* 5 pages, 8 figures, published in Proceedings of the 16th Virtual International Meeting on Fully 3D Image Reconstruction in Radiology and Nuclear Medicine 2021 arXiv:2110.04143
Software Implementation of the Krylov Methods Based Reconstruction for the 3D Cone Beam CT Operator
Oct 26, 2021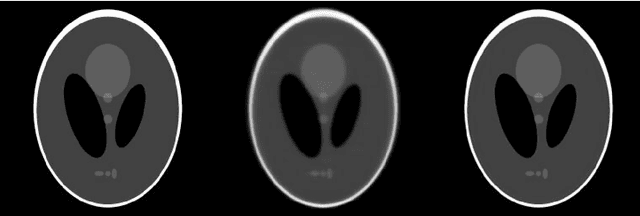
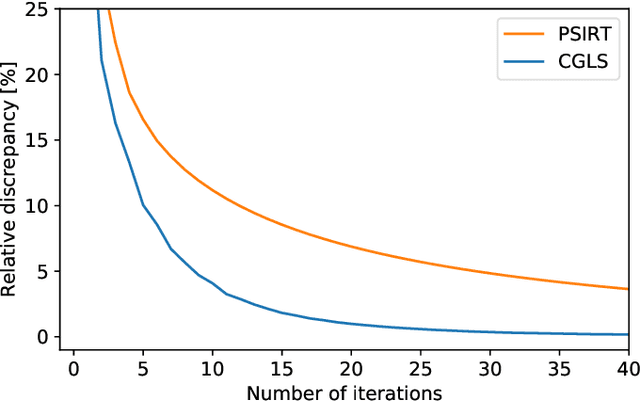
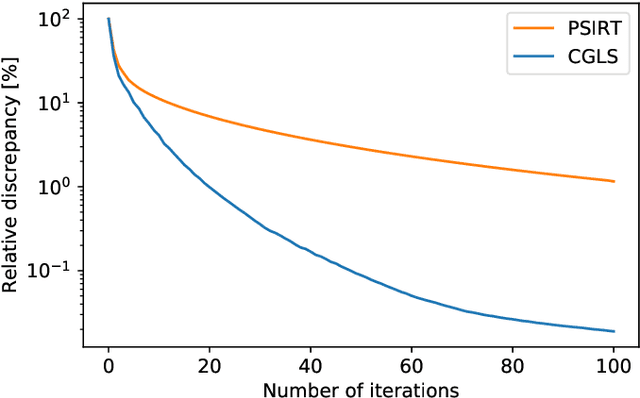
Abstract:Krylov subspace methods are considered a standard tool to solve large systems of linear algebraic equations in many scientific disciplines such as image restoration or solving partial differential equations in mechanics of continuum. In the context of computer tomography however, the mostly used algebraic reconstruction techniques are based on classical iterative schemes. In this work we present software package that implements fully 3D cone beam projection operator and uses Krylov subspace methods, namely CGLS and LSQR to solve related tomographic reconstruction problems. It also implements basic preconditioning strategies. On the example of the cone beam CT reconstruction of 3D Shepp-Logan phantom we show that the speed of convergence of the CGLS clearly outperforms PSIRT algorithm. Therefore Krylov subspace methods present an interesting option for the reconstruction of large 3D cone beam CT problems.
* 5 pages, 3 figures, published in Proceedings of the 16th Virtual International Meeting on Fully 3D Image Reconstruction in Radiology and Nuclear Medicine 2021 arXiv:2110.04143
Cutting Voxel Projector a New Approach to Construct 3D Cone Beam CT Operator
Oct 19, 2021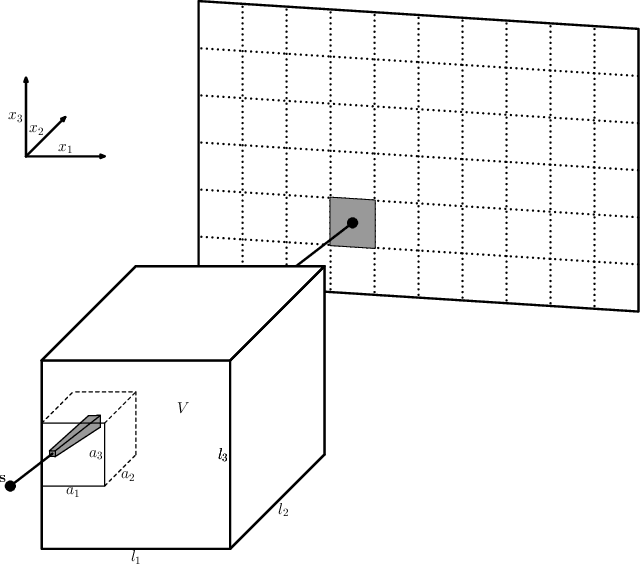
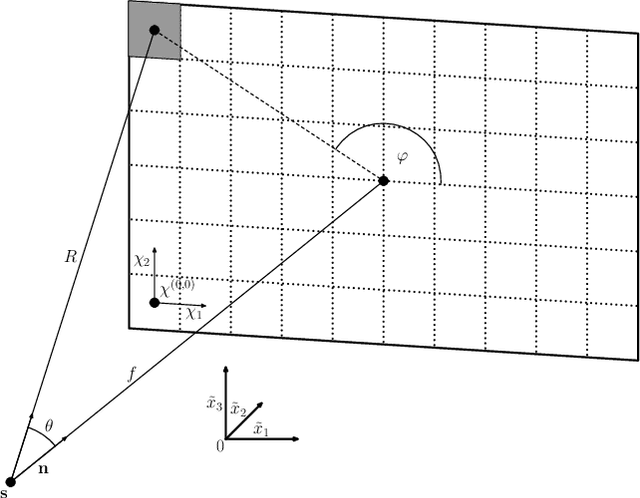
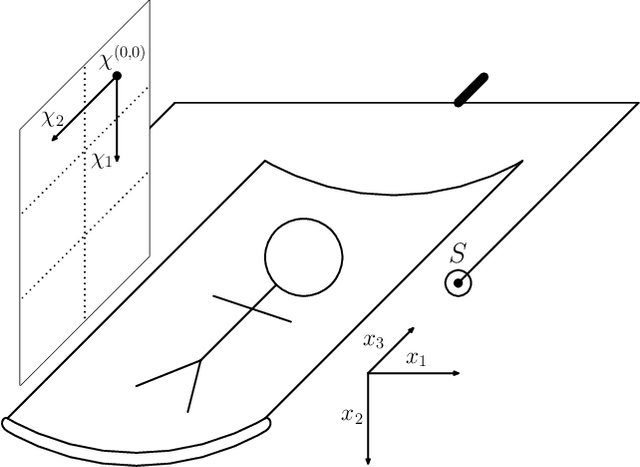
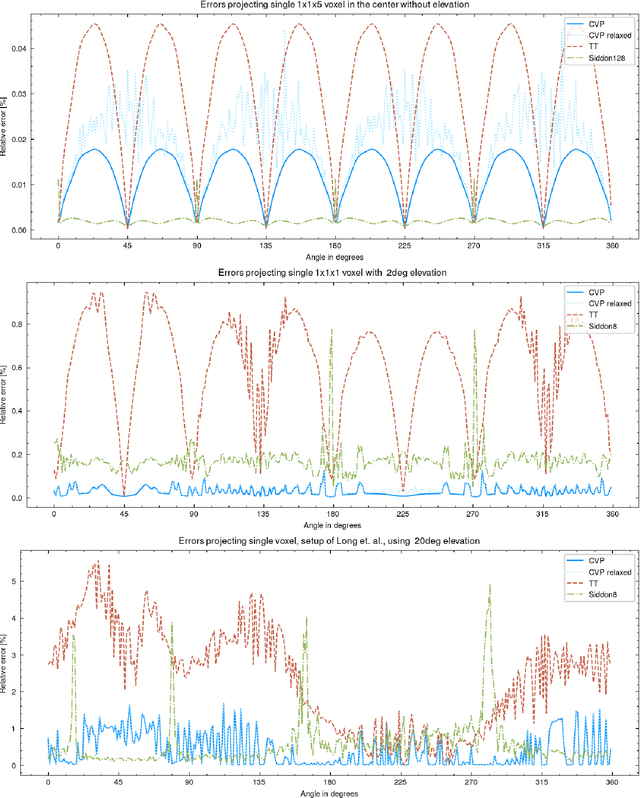
Abstract:In this paper, we introduce a new class of projectors for 3D cone beam tomographic reconstruction. We find analytical formulas for the relationship between the voxel volume projected onto a given detector pixel and its contribution to the extinction value detected on that pixel. Using this approach, we construct a near-exact projector and backprojector that can be used especially for algebraic reconstruction techniques. We have implemented this cutting voxel projector and a less accurate, speed-optimized version of it together with two established projectors, a ray tracing projector based on Siddon's algorithm and a TT footprint projector. We show that the cutting voxel projector achieves, especially for large cone beam angles, noticeably higher accuracy than the TT projector. Moreover, our implementation of the relaxed version of the cutting voxel projector is significantly faster than current footprint projector implementations. We further show that Siddon's algorithm with comparable accuracy would be much slower than the cutting voxel projector. All algorithms are implemented within an open source framework for algebraic reconstruction in OpenCL 1.2 and C++ and are optimized for GPU computation. They are published as open-source software under the GNU GPL 3 license, see https://github.com/kulvait/KCT_cbct.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge