Frank Wacker
Institute of Diagnostic and Interventional Radiology, Hannover Medical School, Hannover, Germany
Liver Segmentation in Time-resolved C-arm CT Volumes Reconstructed from Dynamic Perfusion Scans using Time Separation Technique
Feb 09, 2023



Abstract:Perfusion imaging is a valuable tool for diagnosing and treatment planning for liver tumours. The time separation technique (TST) has been successfully used for modelling C-arm cone-beam computed tomography (CBCT) perfusion data. The reconstruction can be accompanied by the segmentation of the liver - for better visualisation and for generating comprehensive perfusion maps. Recently introduced Turbolift learning has been seen to perform well while working with TST reconstructions, but has not been explored for the time-resolved volumes (TRV) estimated out of TST reconstructions. The segmentation of the TRVs can be useful for tracking the movement of the liver over time. This research explores this possibility by training the multi-scale attention UNet of Turbolift learning at its third stage on the TRVs and shows the robustness of Turbolift learning since it can even work efficiently with the TRVs, resulting in a Dice score of 0.864$\pm$0.004.
Liver Segmentation using Turbolift Learning for CT and Cone-beam C-arm Perfusion Imaging
Jul 20, 2022
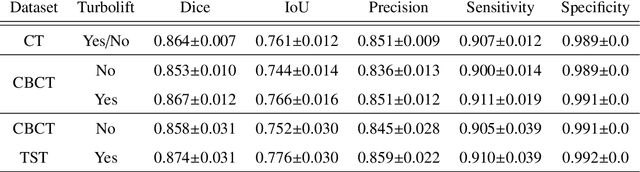
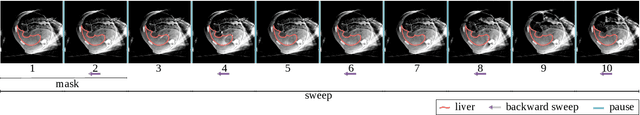
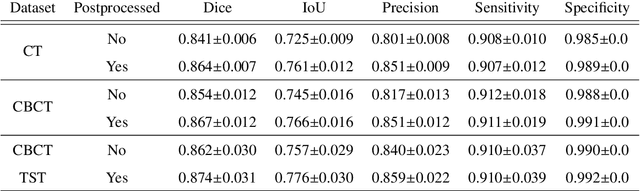
Abstract:Model-based reconstruction employing the time separation technique (TST) was found to improve dynamic perfusion imaging of the liver using C-arm cone-beam computed tomography (CBCT). To apply TST using prior knowledge extracted from CT perfusion data, the liver should be accurately segmented from the CT scans. Reconstructions of primary and model-based CBCT data need to be segmented for proper visualisation and interpretation of perfusion maps. This research proposes Turbolift learning, which trains a modified version of the multi-scale Attention UNet on different liver segmentation tasks serially, following the order of the trainings CT, CBCT, CBCT TST - making the previous trainings act as pre-training stages for the subsequent ones - addressing the problem of limited number of datasets for training. For the final task of liver segmentation from CBCT TST, the proposed method achieved an overall Dice scores of 0.874$\pm$0.031 and 0.905$\pm$0.007 in 6-fold and 4-fold cross-validation experiments, respectively - securing statistically significant improvements over the model, which was trained only for that task. Experiments revealed that Turbolift not only improves the overall performance of the model but also makes it robust against artefacts originating from the embolisation materials and truncation artefacts. Additionally, in-depth analyses confirmed the order of the segmentation tasks. This paper shows the potential of segmenting the liver from CT, CBCT, and CBCT TST, learning from the available limited training data, which can possibly be used in the future for the visualisation and evaluation of the perfusion maps for the treatment evaluation of liver diseases.
Application of Time Separation Technique to Enhance C-arm CT Dynamic Liver Perfusion Imaging
Oct 27, 2021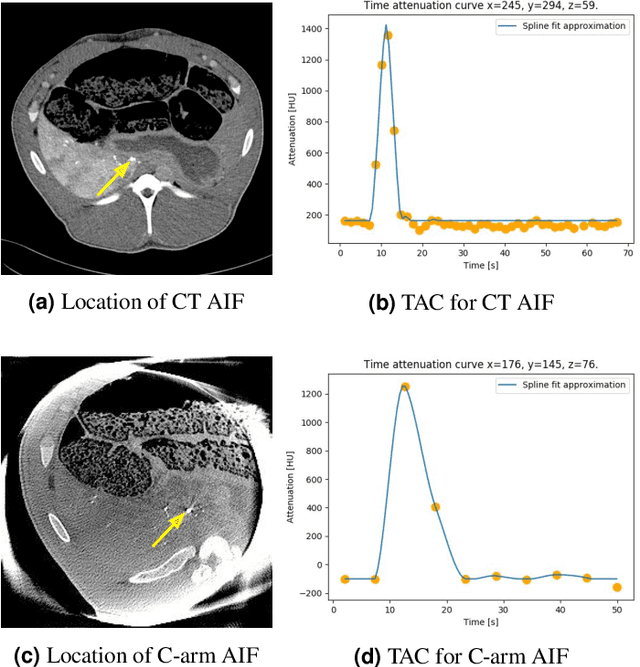
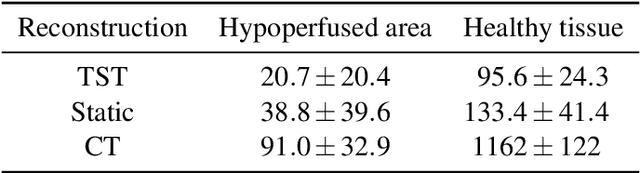
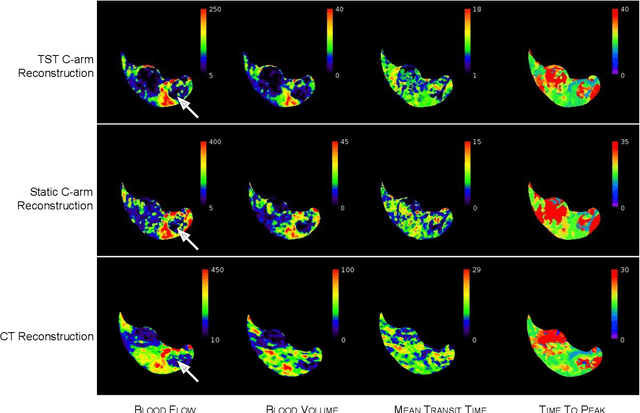
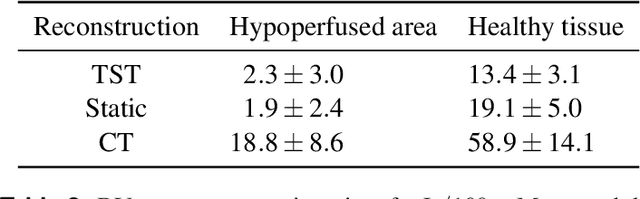
Abstract:Perfusion imaging is an interesting new modality for evaluation and assessment of the liver cancer treatment. C-Arm CT provides a possibility to perform perfusion imaging scans intra-operatively for even faster evaluation. The slow speed of the C-Arm CT rotation and the presence of the noise, however, have an impact on the reconstruction and therefore model based approaches have to be applied. In this work we apply the Time separation technique (TST), to denoise data, speed up reconstruction and improve resulting perfusion images. We show on animal experiment data that Dynamic C-Arm CT Liver Perfusion Imaging together with the processing of the data based on the TST provides comparable results to standard CT liver perfusion imaging.
* 5 pages, 8 figures, published in Proceedings of the 16th Virtual International Meeting on Fully 3D Image Reconstruction in Radiology and Nuclear Medicine 2021 arXiv:2110.04143
2.5D Thermometry Maps for MRI-guided Tumor Ablation
Aug 12, 2021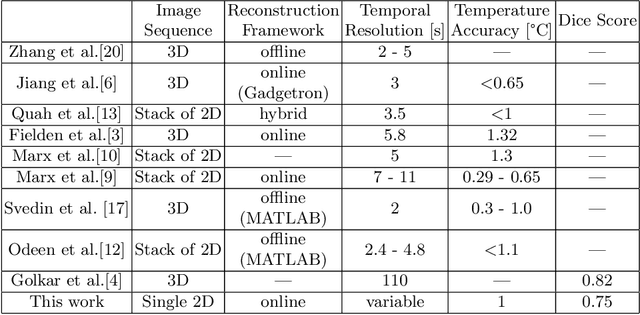
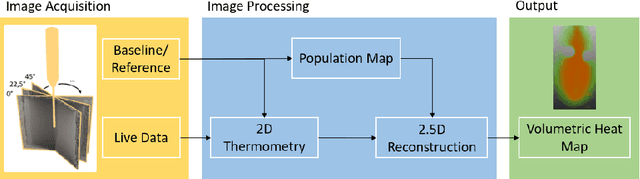
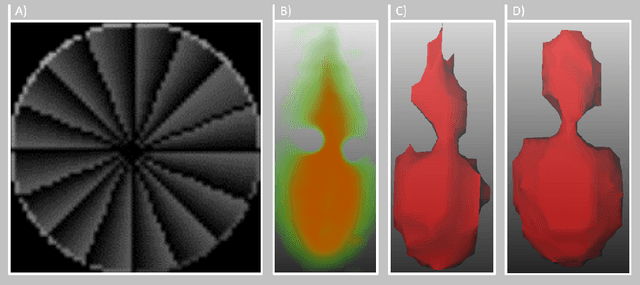
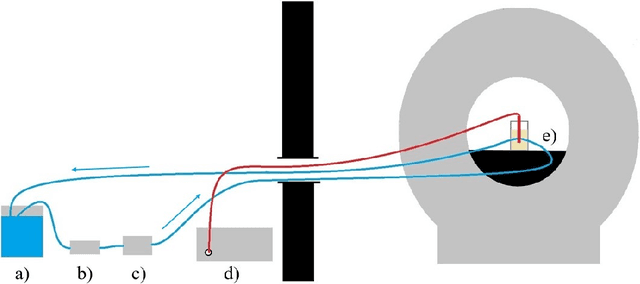
Abstract:Fast and reliable monitoring of volumetric heat distribution during MRI-guided tumor ablation is an urgent clinical need. In this work, we introduce a method for generating 2.5D thermometry maps from uniformly distributed 2D MRI phase images rotated around the applicator's main axis. The images can be fetched directly from the MR device, reducing the delay between image acquisition and visualization. For reconstruction, we use a weighted interpolation on a cylindric coordinate representation to calculate the heat value of voxels in a region of interest. A pilot study on 13 ex vivo bio protein phantoms with flexible tubes to simulate a heat sink effect was conducted to evaluate our method. After thermal ablation, we compared the measured coagulation zone extracted from the post-treatment MR data set with the output of the 2.5D thermometry map. The results show a mean Dice score of 0.75+-0.07, a sensitivity of 0.77+-0.03, and a reconstruction time within 18.02ms+-5.91ms. Future steps should address improving temporal resolution and accuracy, e.g., incorporating advanced bioheat transfer simulations.
$ν$-net: Deep Learning for Generalized Biventricular Cardiac Mass and Function Parameters
Jun 14, 2017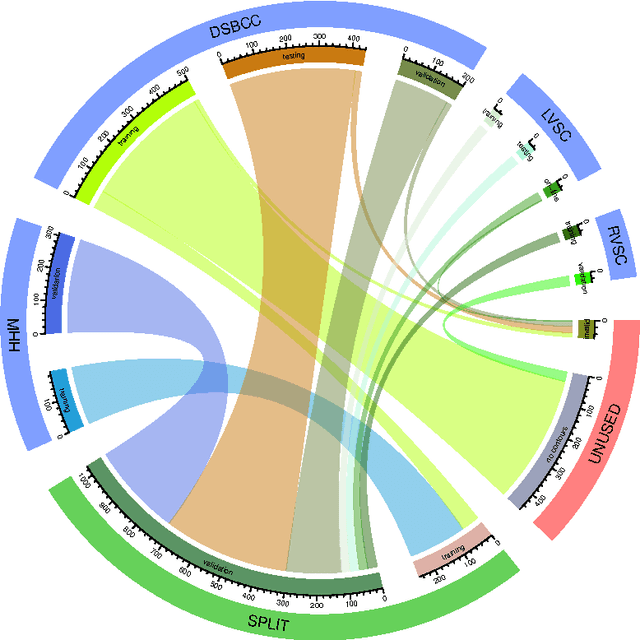

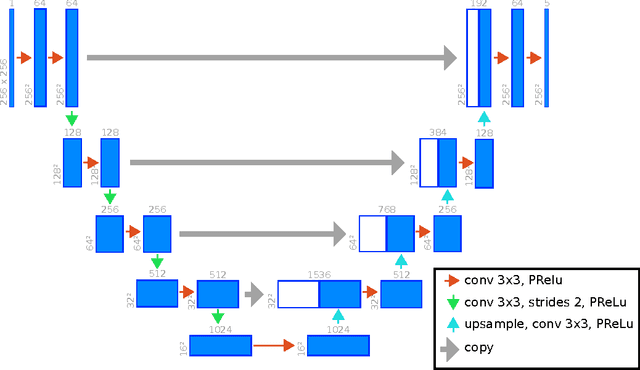
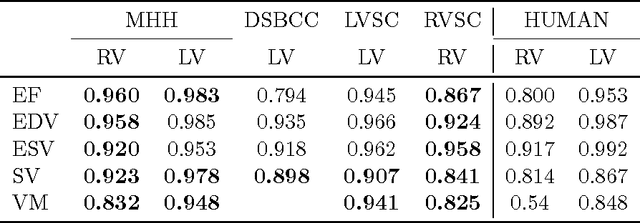
Abstract:Background: Cardiac MRI derived biventricular mass and function parameters, such as end-systolic volume (ESV), end-diastolic volume (EDV), ejection fraction (EF), stroke volume (SV), and ventricular mass (VM) are clinically well established. Image segmentation can be challenging and time-consuming, due to the complex anatomy of the human heart. Objectives: This study introduces $\nu$-net (/nju:n$\varepsilon$t/) -- a deep learning approach allowing for fully-automated high quality segmentation of right (RV) and left ventricular (LV) endocardium and epicardium for extraction of cardiac function parameters. Methods: A set consisting of 253 manually segmented cases has been used to train a deep neural network. Subsequently, the network has been evaluated on 4 different multicenter data sets with a total of over 1000 cases. Results: For LV EF the intraclass correlation coefficient (ICC) is 98, 95, and 80 % (95 %), and for RV EF 96, and 87 % (80 %) on the respective data sets (human expert ICCs reported in parenthesis). The LV VM ICC is 95, and 94 % (84 %), and the RV VM ICC is 83, and 83 % (54 %). This study proposes a simple adjustment procedure, allowing for the adaptation to distinct segmentation philosophies. $\nu$-net exhibits state of-the-art performance in terms of dice coefficient. Conclusions: Biventricular mass and function parameters can be determined reliably in high quality by applying a deep neural network for cardiac MRI segmentation, especially in the anatomically complex right ventricle. Adaption to individual segmentation styles by applying a simple adjustment procedure is viable, allowing for the processing of novel data without time-consuming additional training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge