Johann Bauersachs
$ν$-net: Deep Learning for Generalized Biventricular Cardiac Mass and Function Parameters
Jun 14, 2017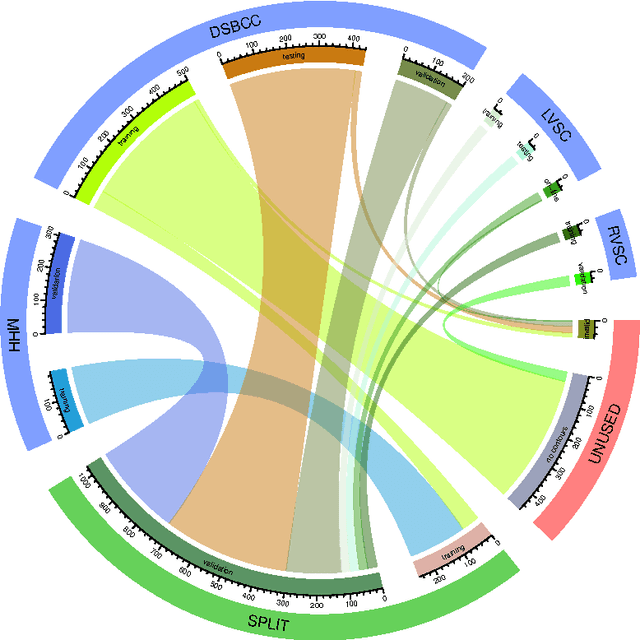

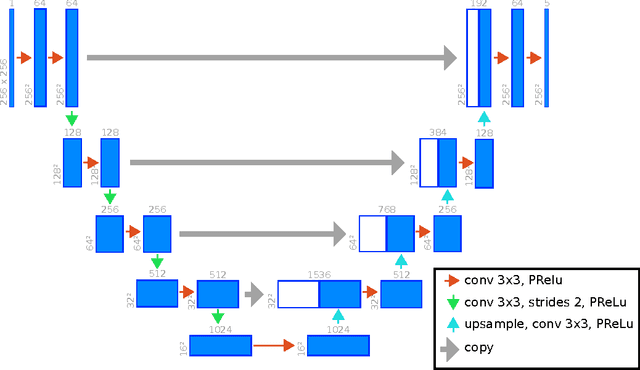
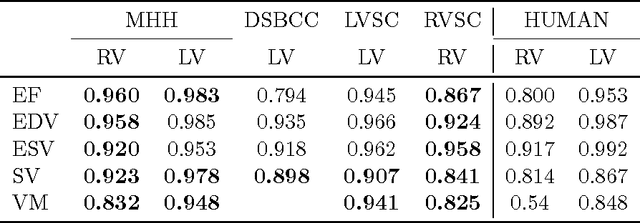
Abstract:Background: Cardiac MRI derived biventricular mass and function parameters, such as end-systolic volume (ESV), end-diastolic volume (EDV), ejection fraction (EF), stroke volume (SV), and ventricular mass (VM) are clinically well established. Image segmentation can be challenging and time-consuming, due to the complex anatomy of the human heart. Objectives: This study introduces $\nu$-net (/nju:n$\varepsilon$t/) -- a deep learning approach allowing for fully-automated high quality segmentation of right (RV) and left ventricular (LV) endocardium and epicardium for extraction of cardiac function parameters. Methods: A set consisting of 253 manually segmented cases has been used to train a deep neural network. Subsequently, the network has been evaluated on 4 different multicenter data sets with a total of over 1000 cases. Results: For LV EF the intraclass correlation coefficient (ICC) is 98, 95, and 80 % (95 %), and for RV EF 96, and 87 % (80 %) on the respective data sets (human expert ICCs reported in parenthesis). The LV VM ICC is 95, and 94 % (84 %), and the RV VM ICC is 83, and 83 % (54 %). This study proposes a simple adjustment procedure, allowing for the adaptation to distinct segmentation philosophies. $\nu$-net exhibits state of-the-art performance in terms of dice coefficient. Conclusions: Biventricular mass and function parameters can be determined reliably in high quality by applying a deep neural network for cardiac MRI segmentation, especially in the anatomically complex right ventricle. Adaption to individual segmentation styles by applying a simple adjustment procedure is viable, allowing for the processing of novel data without time-consuming additional training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge