Gabriele Pergola
When in Doubt, Consult: Expert Debate for Sexism Detection via Confidence-Based Routing
Jan 07, 2026Abstract:Online sexism increasingly appears in subtle, context-dependent forms that evade traditional detection methods. Its interpretation often depends on overlapping linguistic, psychological, legal, and cultural dimensions, which produce mixed and sometimes contradictory signals in annotated datasets. These inconsistencies, combined with label scarcity and class imbalance, result in unstable decision boundaries and cause fine-tuned models to overlook subtler, underrepresented forms of harm. To address these challenges, we propose a two-stage framework that unifies (i) targeted training procedures to better regularize supervision to scarce and noisy data with (ii) selective, reasoning-based inference to handle ambiguous or borderline cases. First, we stabilize the training combining class-balanced focal loss, class-aware batching, and post-hoc threshold calibration, strategies for the firs time adapted for this domain to mitigate label imbalance and noisy supervision. Second, we bridge the gap between efficiency and reasoning with a a dynamic routing mechanism that distinguishes between unambiguous instances and complex cases requiring a deliberative process. This reasoning process results in the novel Collaborative Expert Judgment (CEJ) module which prompts multiple personas and consolidates their reasoning through a judge model. Our approach outperforms existing approaches across several public benchmarks, with F1 gains of +4.48% and +1.30% on EDOS Tasks A and B, respectively, and a +2.79% improvement in ICM on EXIST 2025 Task 1.1.
When in Doubt, Deliberate: Confidence-Based Routing to Expert Debate for Sexism Detection
Dec 21, 2025Abstract:Sexist content online increasingly appears in subtle, context-dependent forms that evade traditional detection methods. Its interpretation often depends on overlapping linguistic, psychological, legal, and cultural dimensions, which produce mixed and sometimes contradictory signals, even in annotated datasets. These inconsistencies, combined with label scarcity and class imbalance, result in unstable decision boundaries and cause fine-tuned models to overlook subtler, underrepresented forms of harm. Together, these limitations point to the need for a design that explicitly addresses the combined effects of (i) underrepresentation, (ii) noise, and (iii) conceptual ambiguity in both data and model predictions. To address these challenges, we propose a two-stage framework that unifies (i) targeted training procedures to adapt supervision to scarce and noisy data with (ii) selective, reasoning-based inference to handle ambiguous or borderline cases. Our training setup applies class-balanced focal loss, class-aware batching, and post-hoc threshold calibration to mitigate label imbalance and noisy supervision. At inference time, a dynamic routing mechanism classifies high-confidence cases directly and escalates uncertain instances to a novel \textit{Collaborative Expert Judgment} (CEJ) module, which prompts multiple personas and consolidates their reasoning through a judge model. Our approach achieves state-of-the-art results across several benchmarks, with a +2.72\% improvement in F1 on the EXIST 2025 Task 1.1, and a gains of +4.48\% and +1.30\% on the EDOS Tasks A and B, respectively.
Explaining Matters: Leveraging Definitions and Semantic Expansion for Sexism Detection
Jun 06, 2025

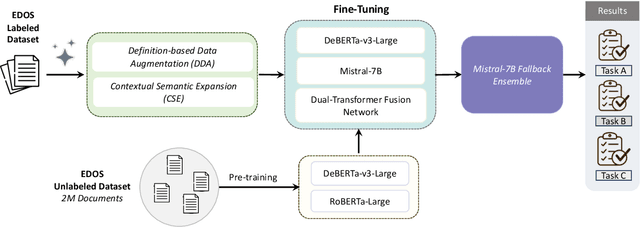

Abstract:The detection of sexism in online content remains an open problem, as harmful language disproportionately affects women and marginalized groups. While automated systems for sexism detection have been developed, they still face two key challenges: data sparsity and the nuanced nature of sexist language. Even in large, well-curated datasets like the Explainable Detection of Online Sexism (EDOS), severe class imbalance hinders model generalization. Additionally, the overlapping and ambiguous boundaries of fine-grained categories introduce substantial annotator disagreement, reflecting the difficulty of interpreting nuanced expressions of sexism. To address these challenges, we propose two prompt-based data augmentation techniques: Definition-based Data Augmentation (DDA), which leverages category-specific definitions to generate semantically-aligned synthetic examples, and Contextual Semantic Expansion (CSE), which targets systematic model errors by enriching examples with task-specific semantic features. To further improve reliability in fine-grained classification, we introduce an ensemble strategy that resolves prediction ties by aggregating complementary perspectives from multiple language models. Our experimental evaluation on the EDOS dataset demonstrates state-of-the-art performance across all tasks, with notable improvements of macro F1 by 1.5 points for binary classification (Task A) and 4.1 points for fine-grained classification (Task C).
SafeSpeech: A Comprehensive and Interactive Tool for Analysing Sexist and Abusive Language in Conversations
Mar 09, 2025Abstract:Detecting toxic language including sexism, harassment and abusive behaviour, remains a critical challenge, particularly in its subtle and context-dependent forms. Existing approaches largely focus on isolated message-level classification, overlooking toxicity that emerges across conversational contexts. To promote and enable future research in this direction, we introduce SafeSpeech, a comprehensive platform for toxic content detection and analysis that bridges message-level and conversation-level insights. The platform integrates fine-tuned classifiers and large language models (LLMs) to enable multi-granularity detection, toxic-aware conversation summarization, and persona profiling. SafeSpeech also incorporates explainability mechanisms, such as perplexity gain analysis, to highlight the linguistic elements driving predictions. Evaluations on benchmark datasets, including EDOS, OffensEval, and HatEval, demonstrate the reproduction of state-of-the-art performance across multiple tasks, including fine-grained sexism detection.
SciGisPy: a Novel Metric for Biomedical Text Simplification via Gist Inference Score
Oct 12, 2024



Abstract:Biomedical literature is often written in highly specialized language, posing significant comprehension challenges for non-experts. Automatic text simplification (ATS) offers a solution by making such texts more accessible while preserving critical information. However, evaluating ATS for biomedical texts is still challenging due to the limitations of existing evaluation metrics. General-domain metrics like SARI, BLEU, and ROUGE focus on surface-level text features, and readability metrics like FKGL and ARI fail to account for domain-specific terminology or assess how well the simplified text conveys core meanings (gist). To address this, we introduce SciGisPy, a novel evaluation metric inspired by Gist Inference Score (GIS) from Fuzzy-Trace Theory (FTT). SciGisPy measures how well a simplified text facilitates the formation of abstract inferences (gist) necessary for comprehension, especially in the biomedical domain. We revise GIS for this purpose by introducing domain-specific enhancements, including semantic chunking, Information Content (IC) theory, and specialized embeddings, while removing unsuitable indexes. Our experimental evaluation on the Cochrane biomedical text simplification dataset demonstrates that SciGisPy outperforms the original GIS formulation, with a significant increase in correctly identified simplified texts (84% versus 44.8%). The results and a thorough ablation study confirm that SciGisPy better captures the essential meaning of biomedical content, outperforming existing approaches.
Society of Medical Simplifiers
Oct 12, 2024



Abstract:Medical text simplification is crucial for making complex biomedical literature more accessible to non-experts. Traditional methods struggle with the specialized terms and jargon of medical texts, lacking the flexibility to adapt the simplification process dynamically. In contrast, recent advancements in large language models (LLMs) present unique opportunities by offering enhanced control over text simplification through iterative refinement and collaboration between specialized agents. In this work, we introduce the Society of Medical Simplifiers, a novel LLM-based framework inspired by the "Society of Mind" (SOM) philosophy. Our approach leverages the strengths of LLMs by assigning five distinct roles, i.e., Layperson, Simplifier, Medical Expert, Language Clarifier, and Redundancy Checker, organized into interaction loops. This structure allows the agents to progressively improve text simplification while maintaining the complexity and accuracy of the original content. Evaluations on the Cochrane text simplification dataset demonstrate that our framework is on par with or outperforms state-of-the-art methods, achieving superior readability and content preservation through controlled simplification processes.
ExDDI: Explaining Drug-Drug Interaction Predictions with Natural Language
Sep 09, 2024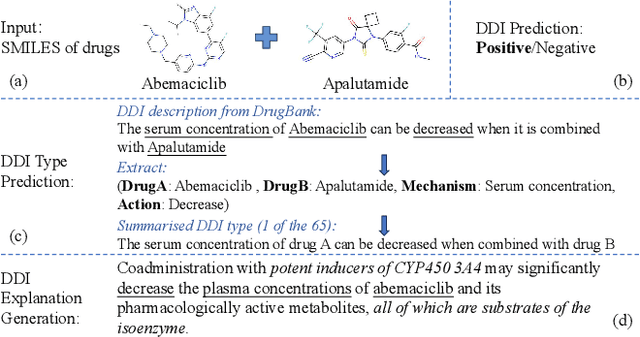


Abstract:Predicting unknown drug-drug interactions (DDIs) is crucial for improving medication safety. Previous efforts in DDI prediction have typically focused on binary classification or predicting DDI categories, with the absence of explanatory insights that could enhance trust in these predictions. In this work, we propose to generate natural language explanations for DDI predictions, enabling the model to reveal the underlying pharmacodynamics and pharmacokinetics mechanisms simultaneously as making the prediction. To do this, we have collected DDI explanations from DDInter and DrugBank and developed various models for extensive experiments and analysis. Our models can provide accurate explanations for unknown DDIs between known drugs. This paper contributes new tools to the field of DDI prediction and lays a solid foundation for further research on generating explanations for DDI predictions.
Cascading Large Language Models for Salient Event Graph Generation
Jun 26, 2024



Abstract:Generating event graphs from long documents is challenging due to the inherent complexity of multiple tasks involved such as detecting events, identifying their relationships, and reconciling unstructured input with structured graphs. Recent studies typically consider all events with equal importance, failing to distinguish salient events crucial for understanding narratives. This paper presents CALLMSAE, a CAscading Large Language Model framework for SAlient Event graph generation, which leverages the capabilities of LLMs and eliminates the need for costly human annotations. We first identify salient events by prompting LLMs to generate summaries, from which salient events are identified. Next, we develop an iterative code refinement prompting strategy to generate event relation graphs, removing hallucinated relations and recovering missing edges. Fine-tuning contextualised graph generation models on the LLM-generated graphs outperforms the models trained on CAEVO-generated data. Experimental results on a human-annotated test set show that the proposed method generates salient and more accurate graphs, outperforming competitive baselines.
DrugWatch: A Comprehensive Multi-Source Data Visualisation Platform for Drug Safety Information
Jun 18, 2024Abstract:Drug safety research is crucial for maintaining public health, often requiring comprehensive data support. However, the resources currently available to the public are limited and fail to provide a comprehensive understanding of the relationship between drugs and their side effects. This paper introduces DrugWatch, an easy-to-use and interactive multi-source information visualisation platform for drug safety study. It allows users to understand common side effects of drugs and their statistical information, flexibly retrieve relevant medical reports, or annotate their own medical texts with our automated annotation tool. Supported by NLP technology and enriched with interactive visual components, we are committed to providing researchers and practitioners with a one-stop information analysis, retrieval, and annotation service. The demonstration video is available at https://www.youtube.com/watch?v=RTqDgxzETjw. We also deployed an online demonstration system at https://drugwatch.net/.
Large Multimodal Model based Standardisation of Pathology Reports with Confidence and their Prognostic Significance
May 03, 2024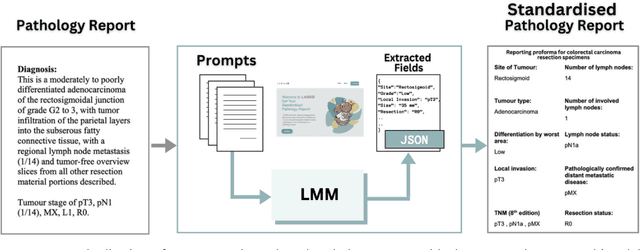
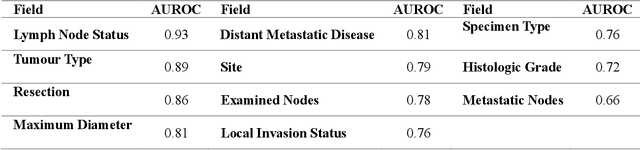
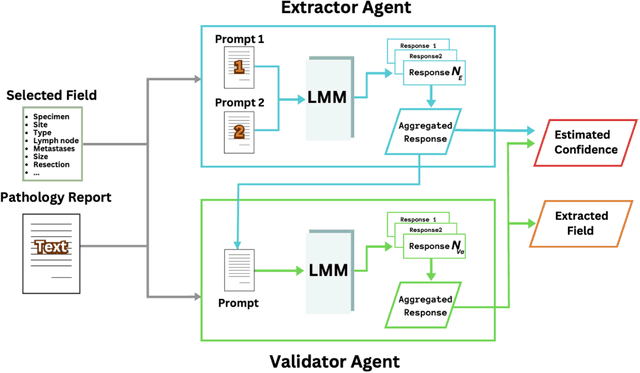
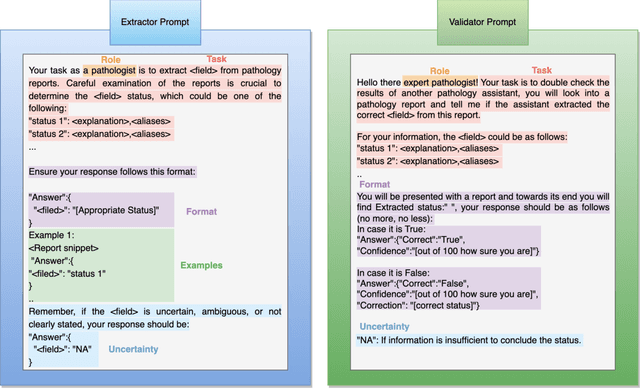
Abstract:Pathology reports are rich in clinical and pathological details but are often presented in free-text format. The unstructured nature of these reports presents a significant challenge limiting the accessibility of their content. In this work, we present a practical approach based on the use of large multimodal models (LMMs) for automatically extracting information from scanned images of pathology reports with the goal of generating a standardised report specifying the value of different fields along with estimated confidence about the accuracy of the extracted fields. The proposed approach overcomes limitations of existing methods which do not assign confidence scores to extracted fields limiting their practical use. The proposed framework uses two stages of prompting a Large Multimodal Model (LMM) for information extraction and validation. The framework generalises to textual reports from multiple medical centres as well as scanned images of legacy pathology reports. We show that the estimated confidence is an effective indicator of the accuracy of the extracted information that can be used to select only accurately extracted fields. We also show the prognostic significance of structured and unstructured data from pathology reports and show that the automatically extracted field values significant prognostic value for patient stratification. The framework is available for evaluation via the URL: https://labieb.dcs.warwick.ac.uk/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge