Fred A. Hamprecht
IWR at Heidelberg University
Equivariance by Local Canonicalization: A Matter of Representation
Sep 30, 2025Abstract:Equivariant neural networks offer strong inductive biases for learning from molecular and geometric data but often rely on specialized, computationally expensive tensor operations. We present a framework to transfers existing tensor field networks into the more efficient local canonicalization paradigm, preserving equivariance while significantly improving the runtime. Within this framework, we systematically compare different equivariant representations in terms of theoretical complexity, empirical runtime, and predictive accuracy. We publish the tensor_frames package, a PyTorchGeometric based implementation for local canonicalization, that enables straightforward integration of equivariance into any standard message passing neural network.
Lorentz Local Canonicalization: How to Make Any Network Lorentz-Equivariant
May 26, 2025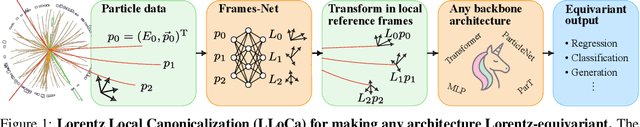
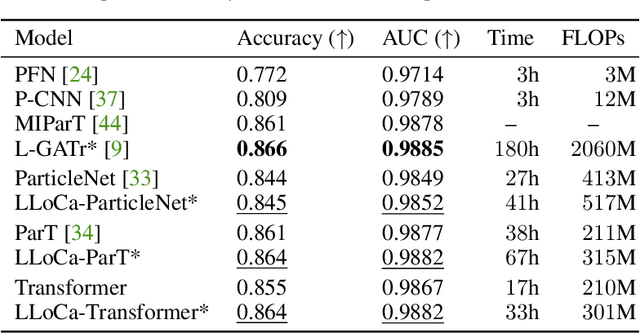
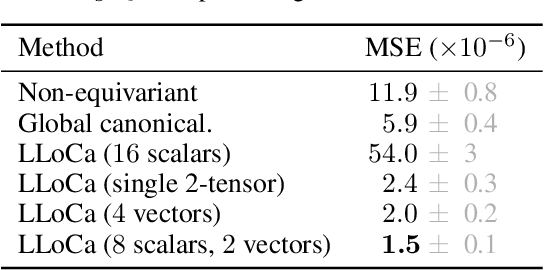
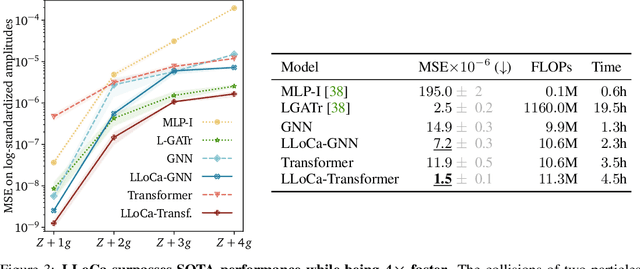
Abstract:Lorentz-equivariant neural networks are becoming the leading architectures for high-energy physics. Current implementations rely on specialized layers, limiting architectural choices. We introduce Lorentz Local Canonicalization (LLoCa), a general framework that renders any backbone network exactly Lorentz-equivariant. Using equivariantly predicted local reference frames, we construct LLoCa-transformers and graph networks. We adapt a recent approach to geometric message passing to the non-compact Lorentz group, allowing propagation of space-time tensorial features. Data augmentation emerges from LLoCa as a special choice of reference frame. Our models surpass state-of-the-art accuracy on relevant particle physics tasks, while being $4\times$ faster and using $5$-$100\times$ fewer FLOPs.
Tensor Frames -- How To Make Any Message Passing Network Equivariant
May 24, 2024



Abstract:In many applications of geometric deep learning, the choice of global coordinate frame is arbitrary, and predictions should be independent of the reference frame. In other words, the network should be equivariant with respect to rotations and reflections of the input, i.e., the transformations of O(d). We present a novel framework for building equivariant message passing architectures and modifying existing non-equivariant architectures to be equivariant. Our approach is based on local coordinate frames, between which geometric information is communicated consistently by including tensorial objects in the messages. Our framework can be applied to message passing on geometric data in arbitrary dimensional Euclidean space. While many other approaches for equivariant message passing require specialized building blocks, such as non-standard normalization layers or non-linearities, our approach can be adapted straightforwardly to any existing architecture without such modifications. We explicitly demonstrate the benefit of O(3)-equivariance for a popular point cloud architecture and produce state-of-the-art results on normal vector regression on point clouds.
SynCellFactory: Generative Data Augmentation for Cell Tracking
Apr 26, 2024Abstract:Cell tracking remains a pivotal yet challenging task in biomedical research. The full potential of deep learning for this purpose is often untapped due to the limited availability of comprehensive and varied training data sets. In this paper, we present SynCellFactory, a generative cell video augmentation. At the heart of SynCellFactory lies the ControlNet architecture, which has been fine-tuned to synthesize cell imagery with photorealistic accuracy in style and motion patterns. This technique enables the creation of synthetic yet realistic cell videos that mirror the complexity of authentic microscopy time-lapses. Our experiments demonstrate that SynCellFactory boosts the performance of well-established deep learning models for cell tracking, particularly when original training data is sparse.
The Central Spanning Tree Problem
Apr 09, 2024Abstract:Spanning trees are an important primitive in many data analysis tasks, when a data set needs to be summarized in terms of its "skeleton", or when a tree-shaped graph over all observations is required for downstream processing. Popular definitions of spanning trees include the minimum spanning tree and the optimum distance spanning tree, a.k.a. the minimum routing cost tree. When searching for the shortest spanning tree but admitting additional branching points, even shorter spanning trees can be realized: Steiner trees. Unfortunately, both minimum spanning and Steiner trees are not robust with respect to noise in the observations; that is, small perturbations of the original data set often lead to drastic changes in the associated spanning trees. In response, we make two contributions when the data lies in a Euclidean space: on the theoretical side, we introduce a new optimization problem, the "(branched) central spanning tree", which subsumes all previously mentioned definitions as special cases. On the practical side, we show empirically that the (branched) central spanning tree is more robust to noise in the data, and as such is better suited to summarize a data set in terms of its skeleton. We also propose a heuristic to address the NP-hard optimization problem, and illustrate its use on single cell RNA expression data from biology and 3D point clouds of plants.
Geometric Autoencoders -- What You See is What You Decode
Jun 30, 2023Abstract:Visualization is a crucial step in exploratory data analysis. One possible approach is to train an autoencoder with low-dimensional latent space. Large network depth and width can help unfolding the data. However, such expressive networks can achieve low reconstruction error even when the latent representation is distorted. To avoid such misleading visualizations, we propose first a differential geometric perspective on the decoder, leading to insightful diagnostics for an embedding's distortion, and second a new regularizer mitigating such distortion. Our ``Geometric Autoencoder'' avoids stretching the embedding spuriously, so that the visualization captures the data structure more faithfully. It also flags areas where little distortion could not be achieved, thus guarding against misinterpretation.
KineticNet: Deep learning a transferable kinetic energy functional for orbital-free density functional theory
May 08, 2023



Abstract:Orbital-free density functional theory (OF-DFT) holds the promise to compute ground state molecular properties at minimal cost. However, it has been held back by our inability to compute the kinetic energy as a functional of the electron density only. We here set out to learn the kinetic energy functional from ground truth provided by the more expensive Kohn-Sham density functional theory. Such learning is confronted with two key challenges: Giving the model sufficient expressivity and spatial context while limiting the memory footprint to afford computations on a GPU; and creating a sufficiently broad distribution of training data to enable iterative density optimization even when starting from a poor initial guess. In response, we introduce KineticNet, an equivariant deep neural network architecture based on point convolutions adapted to the prediction of quantities on molecular quadrature grids. Important contributions include convolution filters with sufficient spatial resolution in the vicinity of the nuclear cusp, an atom-centric sparse but expressive architecture that relays information across multiple bond lengths; and a new strategy to generate varied training data by finding ground state densities in the face of perturbations by a random external potential. KineticNet achieves, for the first time, chemical accuracy of the learned functionals across input densities and geometries of tiny molecules. For two electron systems, we additionally demonstrate OF-DFT density optimization with chemical accuracy.
Theory and Approximate Solvers for Branched Optimal Transport with Multiple Sources
Oct 14, 2022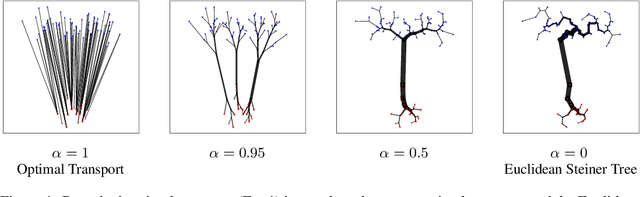


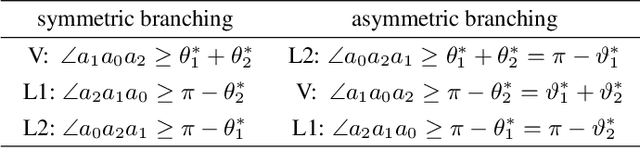
Abstract:Branched Optimal Transport (BOT) is a generalization of optimal transport in which transportation costs along an edge are subadditive. This subadditivity models an increase in transport efficiency when shipping mass along the same route, favoring branched transportation networks. We here study the NP-hard optimization of BOT networks connecting a finite number of sources and sinks in $\mathbb{R}^2$. First, we show how to efficiently find the best geometry of a BOT network for many sources and sinks, given a topology. Second, we argue that a topology with more than three edges meeting at a branching point is never optimal. Third, we show that the results obtained for the Euclidean plane generalize directly to optimal transportation networks on two-dimensional Riemannian manifolds. Finally, we present a simple but effective approximate BOT solver combining geometric optimization with a combinatorial optimization of the network topology.
Contrastive learning unifies $t$-SNE and UMAP
Jun 03, 2022



Abstract:Neighbor embedding methods $t$-SNE and UMAP are the de facto standard for visualizing high-dimensional datasets. They appear to use very different loss functions with different motivations, and the exact relationship between them has been unclear. Here we show that UMAP is effectively negative sampling applied to the $t$-SNE loss function. We explain the difference between negative sampling and noise-contrastive estimation (NCE), which has been used to optimize $t$-SNE under the name NCVis. We prove that, unlike NCE, negative sampling learns a scaled data distribution. When applied in the neighbor embedding setting, it yields more compact embeddings with increased attraction, explaining differences in appearance between UMAP and $t$-SNE. Further, we generalize the notion of negative sampling and obtain a spectrum of embeddings, encompassing visualizations similar to $t$-SNE, NCVis, and UMAP. Finally, we explore the connection between representation learning in the SimCLR setting and neighbor embeddings, and show that (i) $t$-SNE can be optimized using the InfoNCE loss and in a parametric setting; (ii) various contrastive losses with only few noise samples can yield competitive performance in the SimCLR setup.
CellTypeGraph: A New Geometric Computer Vision Benchmark
May 17, 2022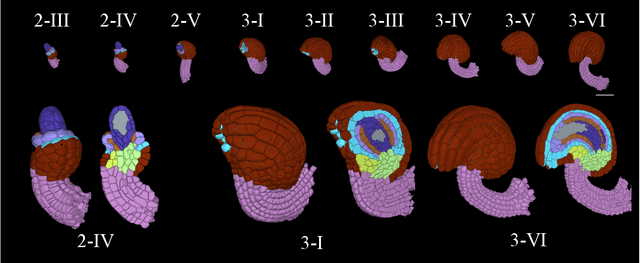
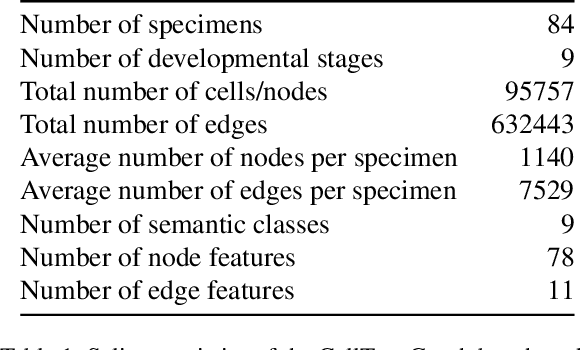
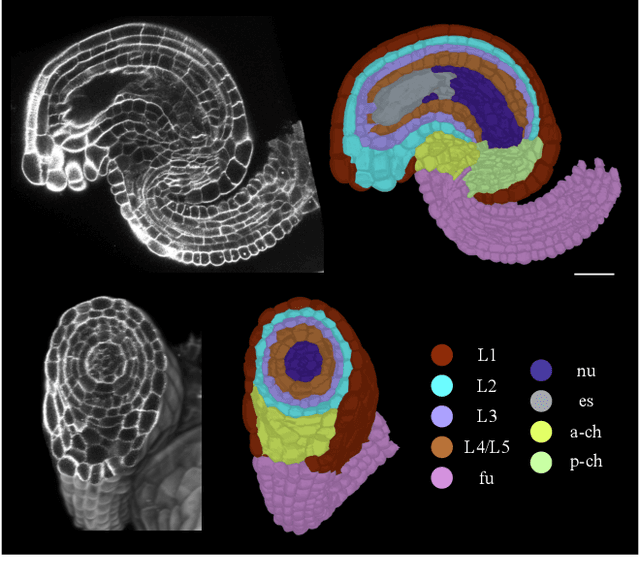
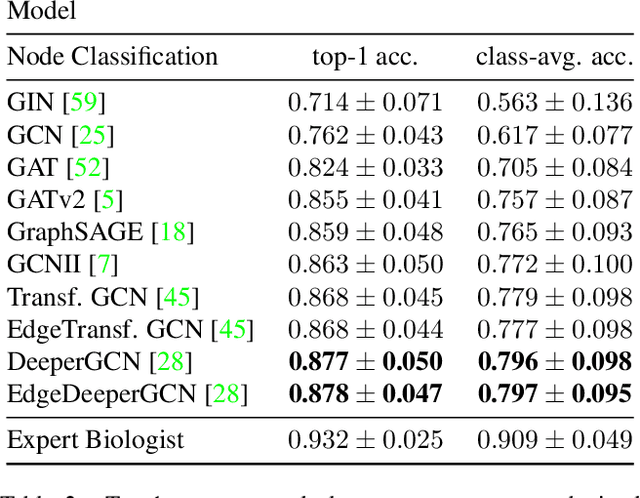
Abstract:Classifying all cells in an organ is a relevant and difficult problem from plant developmental biology. We here abstract the problem into a new benchmark for node classification in a geo-referenced graph. Solving it requires learning the spatial layout of the organ including symmetries. To allow the convenient testing of new geometrical learning methods, the benchmark of Arabidopsis thaliana ovules is made available as a PyTorch data loader, along with a large number of precomputed features. Finally, we benchmark eight recent graph neural network architectures, finding that DeeperGCN currently works best on this problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge