Francesca Ieva
Interpretable phenotyping of Heart Failure patients with Dutch discharge letters
May 30, 2025Abstract:Objective: Heart failure (HF) patients present with diverse phenotypes affecting treatment and prognosis. This study evaluates models for phenotyping HF patients based on left ventricular ejection fraction (LVEF) classes, using structured and unstructured data, assessing performance and interpretability. Materials and Methods: The study analyzes all HF hospitalizations at both Amsterdam UMC hospitals (AMC and VUmc) from 2015 to 2023 (33,105 hospitalizations, 16,334 patients). Data from AMC were used for model training, and from VUmc for external validation. The dataset was unlabelled and included tabular clinical measurements and discharge letters. Silver labels for LVEF classes were generated by combining diagnosis codes, echocardiography results, and textual mentions. Gold labels were manually annotated for 300 patients for testing. Multiple Transformer-based (black-box) and Aug-Linear (white-box) models were trained and compared with baselines on structured and unstructured data. To evaluate interpretability, two clinicians annotated 20 discharge letters by highlighting information they considered relevant for LVEF classification. These were compared to SHAP and LIME explanations from black-box models and the inherent explanations of Aug-Linear models. Results: BERT-based and Aug-Linear models, using discharge letters alone, achieved the highest classification results (AUC=0.84 for BERT, 0.81 for Aug-Linear on external validation), outperforming baselines. Aug-Linear explanations aligned more closely with clinicians' explanations than post-hoc explanations on black-box models. Conclusions: Discharge letters emerged as the most informative source for phenotyping HF patients. Aug-Linear models matched black-box performance while providing clinician-aligned interpretability, supporting their use in transparent clinical decision-making.
DISARM++: Beyond scanner-free harmonization
May 06, 2025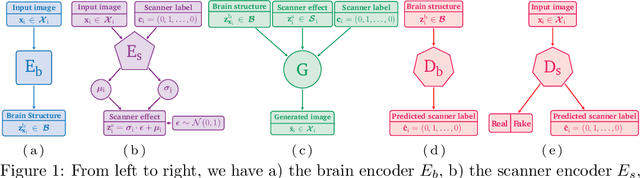
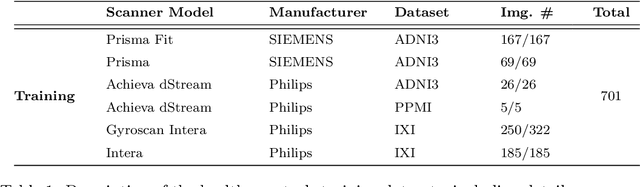
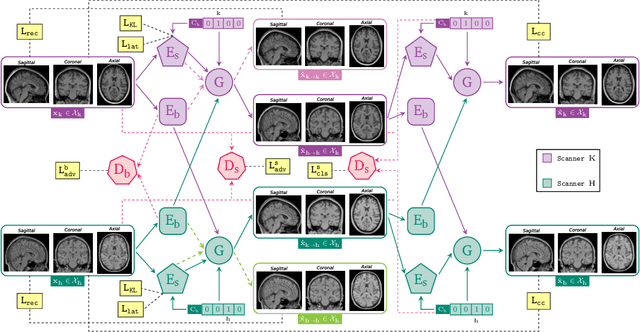
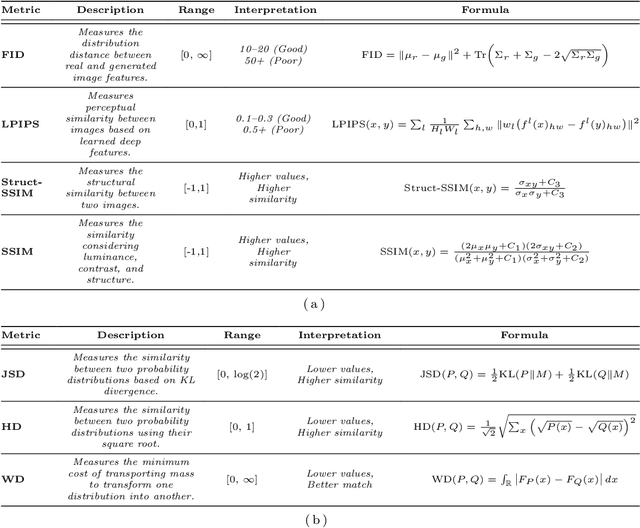
Abstract:Harmonization of T1-weighted MR images across different scanners is crucial for ensuring consistency in neuroimaging studies. This study introduces a novel approach to direct image harmonization, moving beyond feature standardization to ensure that extracted features remain inherently reliable for downstream analysis. Our method enables image transfer in two ways: (1) mapping images to a scanner-free space for uniform appearance across all scanners, and (2) transforming images into the domain of a specific scanner used in model training, embedding its unique characteristics. Our approach presents strong generalization capability, even for unseen scanners not included in the training phase. We validated our method using MR images from diverse cohorts, including healthy controls, traveling subjects, and individuals with Alzheimer's disease (AD). The model's effectiveness is tested in multiple applications, such as brain age prediction (R2 = 0.60 \pm 0.05), biomarker extraction, AD classification (Test Accuracy = 0.86 \pm 0.03), and diagnosis prediction (AUC = 0.95). In all cases, our harmonization technique outperforms state-of-the-art methods, showing improvements in both reliability and predictive accuracy. Moreover, our approach eliminates the need for extensive preprocessing steps, such as skull-stripping, which can introduce errors by misclassifying brain and non-brain structures. This makes our method particularly suitable for applications that require full-head analysis, including research on head trauma and cranial deformities. Additionally, our harmonization model does not require retraining for new datasets, allowing smooth integration into various neuroimaging workflows. By ensuring scanner-invariant image quality, our approach provides a robust and efficient solution for improving neuroimaging studies across diverse settings. The code is available at this link.
NLP-based assessment of prescription appropriateness from Italian referrals
Jan 24, 2025Abstract:Objective: This study proposes a Natural Language Processing pipeline to evaluate prescription appropriateness in Italian referrals, where reasons for prescriptions are recorded only as free text, complicating automated comparisons with guidelines. The pipeline aims to derive, for the first time, a comprehensive summary of the reasons behind these referrals and a quantification of their appropriateness. While demonstrated in a specific case study, the approach is designed to generalize to other types of examinations. Methods: Leveraging embeddings from a transformer-based model, the proposed approach clusters referral texts, maps clusters to labels, and aligns these labels with existing guidelines. We present a case study on a dataset of 496,971 referrals, consisting of all referrals for venous echocolordopplers of the lower limbs between 2019 and 2021 in the Lombardy Region. A sample of 1,000 referrals was manually annotated to validate the results. Results: The pipeline exhibited high performance for referrals' reasons (Prec=92.43%, Rec=83.28%) and excellent results for referrals' appropriateness (Prec=93.58%, Rec=91.52%) on the annotated subset. Analysis of the entire dataset identified clusters matching guideline-defined reasons - both appropriate and inappropriate - as well as clusters not addressed in the guidelines. Overall, 34.32% of referrals were marked as appropriate, 34.07% inappropriate, 14.37% likely inappropriate, and 17.24% could not be mapped to guidelines. Conclusions: The proposed pipeline effectively assessed prescription appropriateness across a large dataset, serving as a valuable tool for health authorities. Findings have informed the Lombardy Region's efforts to strengthen recommendations and reduce the burden of inappropriate referrals.
Weakly-supervised diagnosis identification from Italian discharge letters
Oct 19, 2024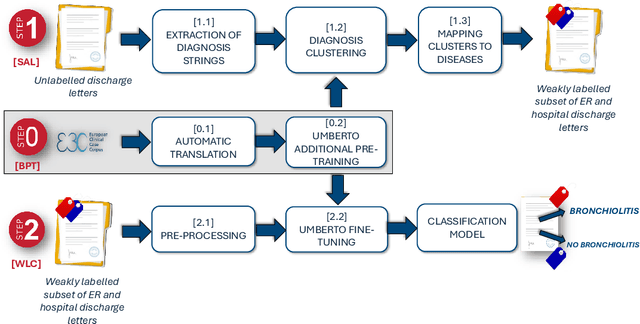
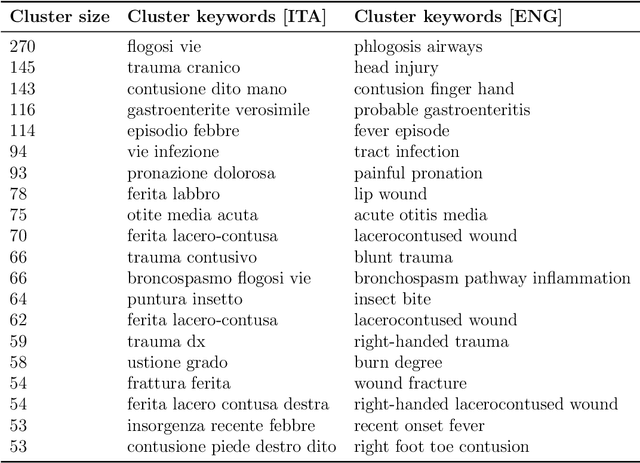
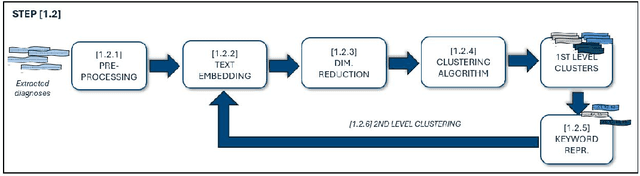

Abstract:Objective: Recognizing diseases from discharge letters is crucial for cohort selection and epidemiological analyses, as this is the only type of data consistently produced across hospitals. This is a classic document classification problem, typically requiring supervised learning. However, manual annotation of large datasets of discharge letters is uncommon since it is extremely time-consuming. We propose a novel weakly-supervised pipeline to recognize diseases from Italian discharge letters. Methods: Our Natural Language Processing pipeline is based on a fine-tuned version of the Italian Umberto model. The pipeline extracts diagnosis-related sentences from a subset of letters and applies a two-level clustering using the embeddings generated by the fine-tuned Umberto model. These clusters are summarized and those mapped to the diseases of interest are selected as weak labels. Finally, the same BERT-based model is trained using these weak labels to detect the targeted diseases. Results: A case study related to the identification of bronchiolitis with 33'176 Italian discharge letters from 44 hospitals in the Veneto Region shows the potential of our method, with an AUC of 77.7 % and an F1-Score of 75.1 % on manually annotated labels, improving compared to other non-supervised methods and with a limited loss compared to fully supervised methods. Results are robust to the cluster selection and the identified clusters highlight the potential to recognize a variety of diseases. Conclusions: This study demonstrates the feasibility of diagnosis identification from Italian discharge letters in the absence of labelled data. Our pipeline showed strong performance and robustness, and its flexibility allows for easy adaptation to various diseases. This approach offers a scalable solution for clinical text classification, reducing the need for manual annotation while maintaining good accuracy.
Scaling Survival Analysis in Healthcare with Federated Survival Forests: A Comparative Study on Heart Failure and Breast Cancer Genomics
Aug 04, 2023Abstract:Survival analysis is a fundamental tool in medicine, modeling the time until an event of interest occurs in a population. However, in real-world applications, survival data are often incomplete, censored, distributed, and confidential, especially in healthcare settings where privacy is critical. The scarcity of data can severely limit the scalability of survival models to distributed applications that rely on large data pools. Federated learning is a promising technique that enables machine learning models to be trained on multiple datasets without compromising user privacy, making it particularly well-suited for addressing the challenges of survival data and large-scale survival applications. Despite significant developments in federated learning for classification and regression, many directions remain unexplored in the context of survival analysis. In this work, we propose an extension of the Federated Survival Forest algorithm, called FedSurF++. This federated ensemble method constructs random survival forests in heterogeneous federations. Specifically, we investigate several new tree sampling methods from client forests and compare the results with state-of-the-art survival models based on neural networks. The key advantage of FedSurF++ is its ability to achieve comparable performance to existing methods while requiring only a single communication round to complete. The extensive empirical investigation results in a significant improvement from the algorithmic and privacy preservation perspectives, making the original FedSurF algorithm more efficient, robust, and private. We also present results on two real-world datasets demonstrating the success of FedSurF++ in real-world healthcare studies. Our results underscore the potential of FedSurF++ to improve the scalability and effectiveness of survival analysis in distributed settings while preserving user privacy.
Imaging-based representation and stratification of intra-tumor Heterogeneity via tree-edit distance
Aug 09, 2022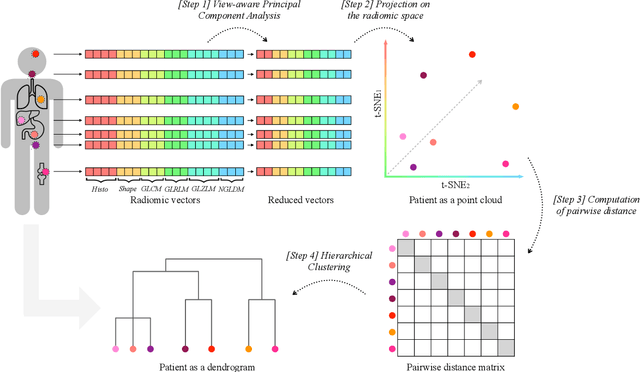
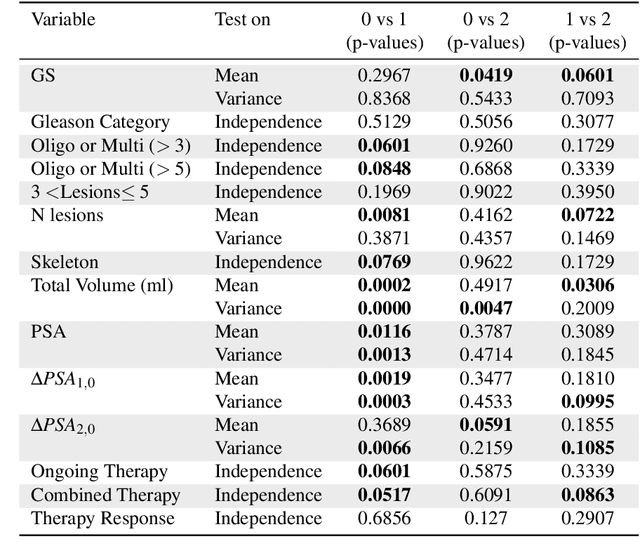
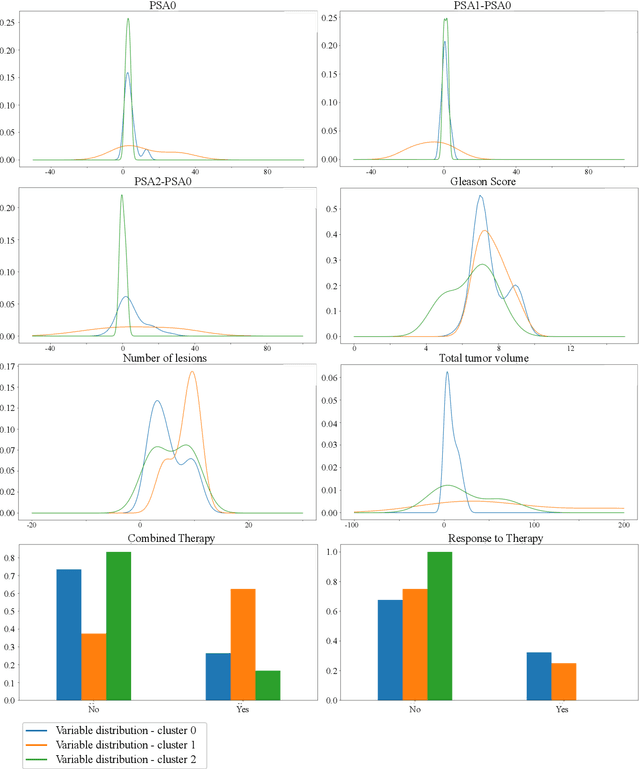
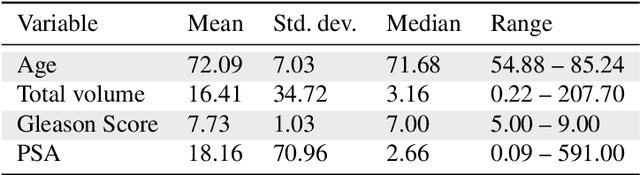
Abstract:Personalized medicine is the future of medical practice. In oncology, tumor heterogeneity assessment represents a pivotal step for effective treatment planning and prognosis prediction. Despite new procedures for DNA sequencing and analysis, non-invasive methods for tumor characterization are needed to impact on daily routine. On purpose, imaging texture analysis is rapidly scaling, holding the promise to surrogate histopathological assessment of tumor lesions. In this work, we propose a tree-based representation strategy for describing intra-tumor heterogeneity of patients affected by metastatic cancer. We leverage radiomics information extracted from PET/CT imaging and we provide an exhaustive and easily readable summary of the disease spreading. We exploit this novel patient representation to perform cancer subtyping according to hierarchical clustering technique. To this purpose, a new heterogeneity-based distance between trees is defined and applied to a case study of Prostate Cancer (PCa). Clusters interpretation is explored in terms of concordance with severity status, tumor burden and biological characteristics. Results are promising, as the proposed method outperforms current literature approaches. Ultimately, the proposed methods draws a general analysis framework that would allow to extract knowledge from daily acquired imaging data of patients and provide insights for effective treatment planning.
Learning Signal Representations for EEG Cross-Subject Channel Selection and Trial Classification
Jun 20, 2021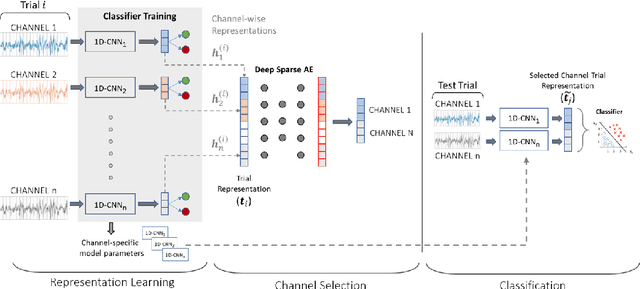
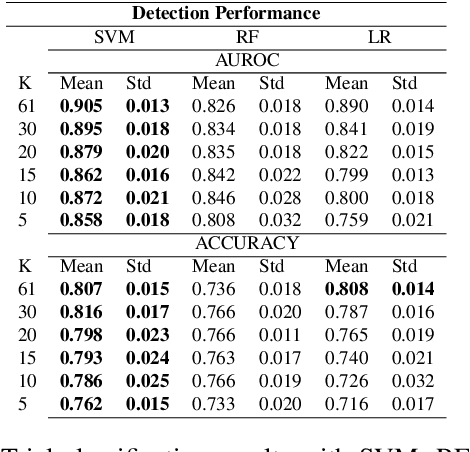
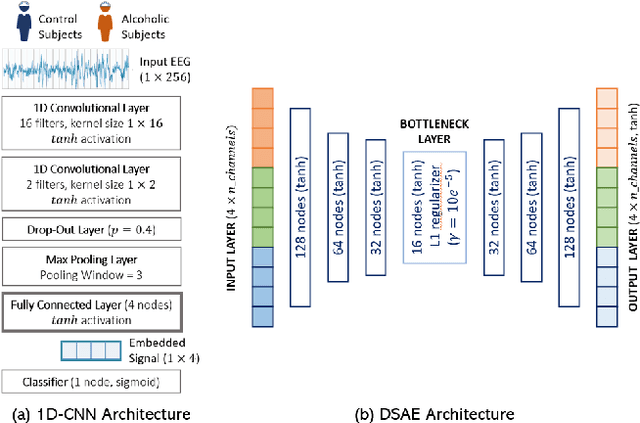
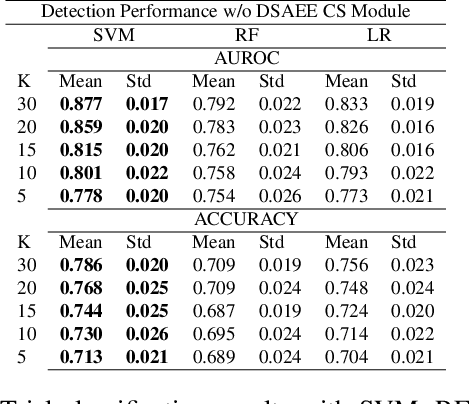
Abstract:EEG technology finds applications in several domains. Currently, most EEG systems require subjects to wear several electrodes on the scalp to be effective. However, several channels might include noisy information, redundant signals, induce longer preparation times and increase computational times of any automated system for EEG decoding. One way to reduce the signal-to-noise ratio and improve classification accuracy is to combine channel selection with feature extraction, but EEG signals are known to present high inter-subject variability. In this work we introduce a novel algorithm for subject-independent channel selection of EEG recordings. Considering multi-channel trial recordings as statistical units and the EEG decoding task as the class of reference, the algorithm (i) exploits channel-specific 1D-Convolutional Neural Networks (1D-CNNs) as feature extractors in a supervised fashion to maximize class separability; (ii) it reduces a high dimensional multi-channel trial representation into a unique trial vector by concatenating the channels' embeddings and (iii) recovers the complex inter-channel relationships during channel selection, by exploiting an ensemble of AutoEncoders (AE) to identify from these vectors the most relevant channels to perform classification. After training, the algorithm can be exploited by transferring only the parametrized subgroup of selected channel-specific 1D-CNNs to new signals from new subjects and obtain low-dimensional and highly informative trial vectors to be fed to any classifier.
Feature Selection for Imbalanced Data with Deep Sparse Autoencoders Ensemble
Mar 22, 2021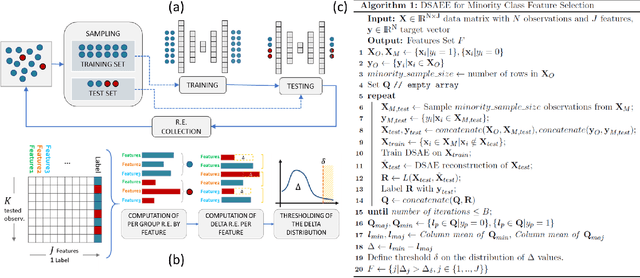
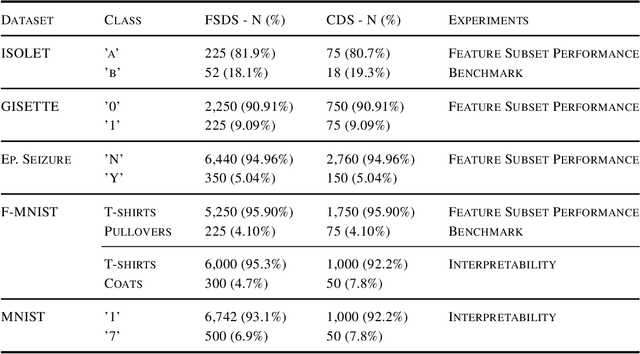

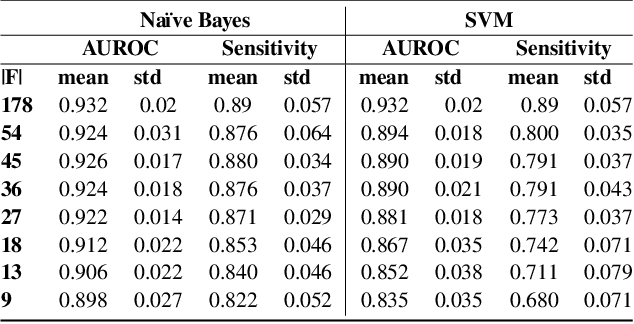
Abstract:Class imbalance is a common issue in many domain applications of learning algorithms. Oftentimes, in the same domains it is much more relevant to correctly classify and profile minority class observations. This need can be addressed by Feature Selection (FS), that offers several further advantages, s.a. decreasing computational costs, aiding inference and interpretability. However, traditional FS techniques may become sub-optimal in the presence of strongly imbalanced data. To achieve FS advantages in this setting, we propose a filtering FS algorithm ranking feature importance on the basis of the Reconstruction Error of a Deep Sparse AutoEncoders Ensemble (DSAEE). We use each DSAE trained only on majority class to reconstruct both classes. From the analysis of the aggregated Reconstruction Error, we determine the features where the minority class presents a different distribution of values w.r.t. the overrepresented one, thus identifying the most relevant features to discriminate between the two. We empirically demonstrate the efficacy of our algorithm in several experiments on high-dimensional datasets of varying sample size, showcasing its capability to select relevant and generalizable features to profile and classify minority class, outperforming other benchmark FS methods. We also briefly present a real application in radiogenomics, where the methodology was applied successfully.
Learning High-Order Interactions via Targeted Pattern Search
Feb 23, 2021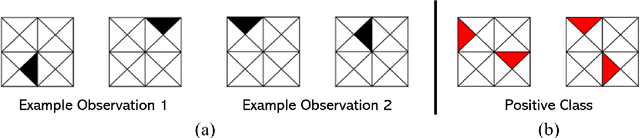
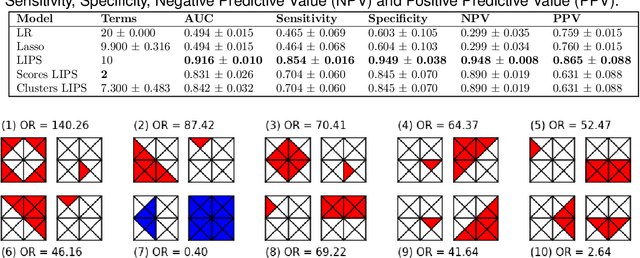
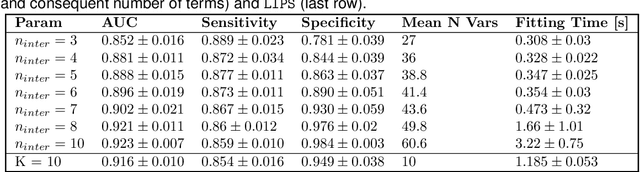
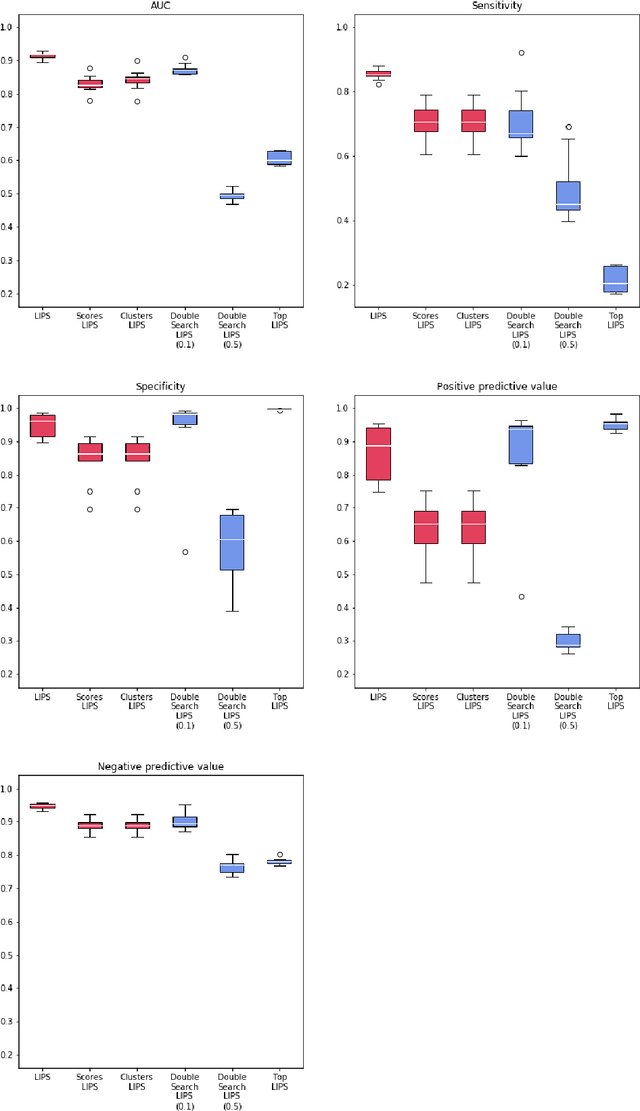
Abstract:Logistic Regression (LR) is a widely used statistical method in empirical binary classification studies. However, real-life scenarios oftentimes share complexities that prevent from the use of the as-is LR model, and instead highlight the need to include high-order interactions to capture data variability. This becomes even more challenging because of: (i) datasets growing wider, with more and more variables; (ii) studies being typically conducted in strongly imbalanced settings; (iii) samples going from very large to extremely small; (iv) the need of providing both predictive models and interpretable results. In this paper we present a novel algorithm, Learning high-order Interactions via targeted Pattern Search (LIPS), to select interaction terms of varying order to include in a LR model for an imbalanced binary classification task when input data are categorical. LIPS's rationale stems from the duality between item sets and categorical interactions. The algorithm relies on an interaction learning step based on a well-known frequent item set mining algorithm, and a novel dissimilarity-based interaction selection step that allows the user to specify the number of interactions to be included in the LR model. In addition, we particularize two variants (Scores LIPS and Clusters LIPS), that can address even more specific needs. Through a set of experiments we validate our algorithm and prove its wide applicability to real-life research scenarios, showing that it outperforms a benchmark state-of-the-art algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge