Michela C. Massi
Learning Signal Representations for EEG Cross-Subject Channel Selection and Trial Classification
Jun 20, 2021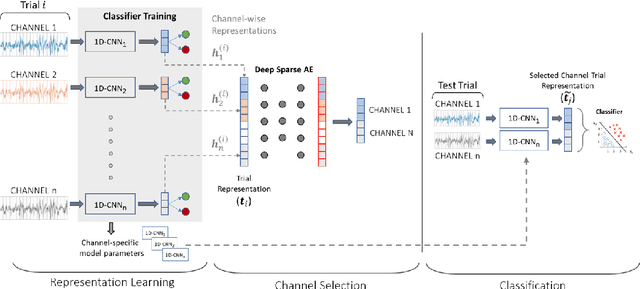
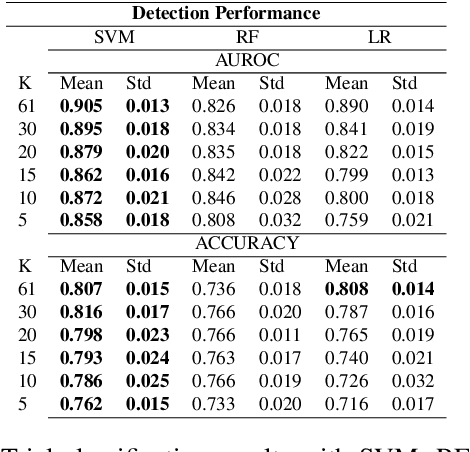
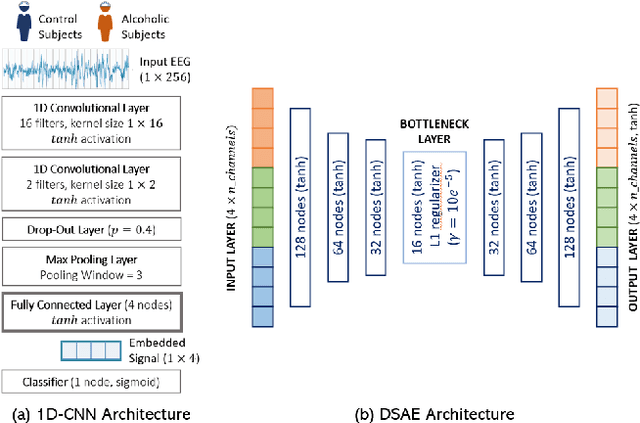
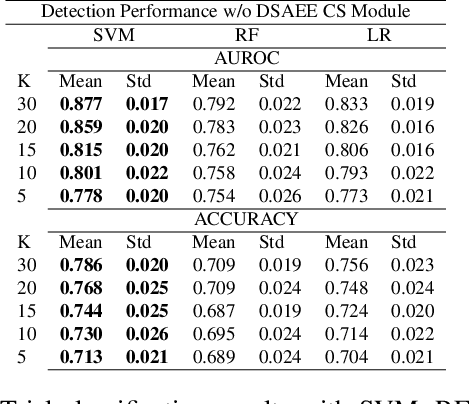
Abstract:EEG technology finds applications in several domains. Currently, most EEG systems require subjects to wear several electrodes on the scalp to be effective. However, several channels might include noisy information, redundant signals, induce longer preparation times and increase computational times of any automated system for EEG decoding. One way to reduce the signal-to-noise ratio and improve classification accuracy is to combine channel selection with feature extraction, but EEG signals are known to present high inter-subject variability. In this work we introduce a novel algorithm for subject-independent channel selection of EEG recordings. Considering multi-channel trial recordings as statistical units and the EEG decoding task as the class of reference, the algorithm (i) exploits channel-specific 1D-Convolutional Neural Networks (1D-CNNs) as feature extractors in a supervised fashion to maximize class separability; (ii) it reduces a high dimensional multi-channel trial representation into a unique trial vector by concatenating the channels' embeddings and (iii) recovers the complex inter-channel relationships during channel selection, by exploiting an ensemble of AutoEncoders (AE) to identify from these vectors the most relevant channels to perform classification. After training, the algorithm can be exploited by transferring only the parametrized subgroup of selected channel-specific 1D-CNNs to new signals from new subjects and obtain low-dimensional and highly informative trial vectors to be fed to any classifier.
A Deep Variational Approach to Clustering Survival Data
Jun 10, 2021



Abstract:Survival analysis has gained significant attention in the medical domain and has many far-reaching applications. Although a variety of machine learning methods have been introduced for tackling time-to-event prediction in unstructured data with complex dependencies, clustering of survival data remains an under-explored problem. The latter is particularly helpful in discovering patient subpopulations whose survival is regulated by different generative mechanisms, a critical problem in precision medicine. To this end, we introduce a novel probabilistic approach to cluster survival data in a variational deep clustering setting. Our proposed method employs a deep generative model to uncover the underlying distribution of both the explanatory variables and the potentially censored survival times. We compare our model to the related work on survival clustering in comprehensive experiments on a range of synthetic, semi-synthetic, and real-world datasets. Our proposed method performs better at identifying clusters and is competitive at predicting survival times in terms of the concordance index and relative absolute error. To further demonstrate the usefulness of our approach, we show that our method identifies meaningful clusters from an observational cohort of hemodialysis patients that are consistent with previous clinical findings.
Feature Selection for Imbalanced Data with Deep Sparse Autoencoders Ensemble
Mar 22, 2021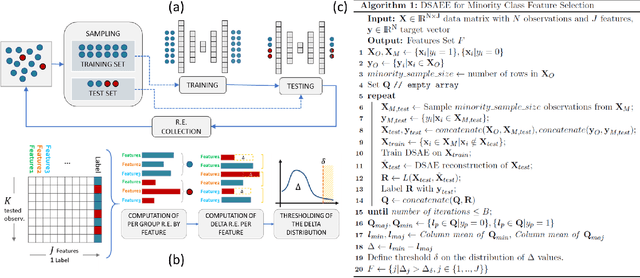
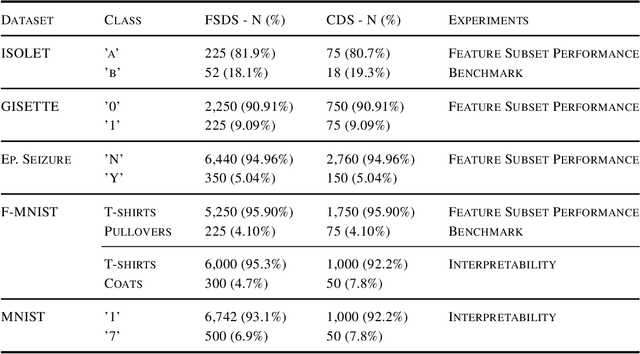

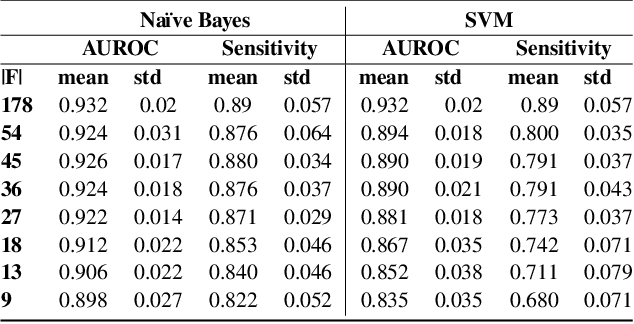
Abstract:Class imbalance is a common issue in many domain applications of learning algorithms. Oftentimes, in the same domains it is much more relevant to correctly classify and profile minority class observations. This need can be addressed by Feature Selection (FS), that offers several further advantages, s.a. decreasing computational costs, aiding inference and interpretability. However, traditional FS techniques may become sub-optimal in the presence of strongly imbalanced data. To achieve FS advantages in this setting, we propose a filtering FS algorithm ranking feature importance on the basis of the Reconstruction Error of a Deep Sparse AutoEncoders Ensemble (DSAEE). We use each DSAE trained only on majority class to reconstruct both classes. From the analysis of the aggregated Reconstruction Error, we determine the features where the minority class presents a different distribution of values w.r.t. the overrepresented one, thus identifying the most relevant features to discriminate between the two. We empirically demonstrate the efficacy of our algorithm in several experiments on high-dimensional datasets of varying sample size, showcasing its capability to select relevant and generalizable features to profile and classify minority class, outperforming other benchmark FS methods. We also briefly present a real application in radiogenomics, where the methodology was applied successfully.
Learning High-Order Interactions via Targeted Pattern Search
Feb 23, 2021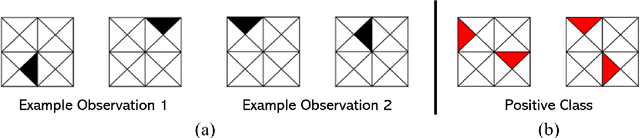
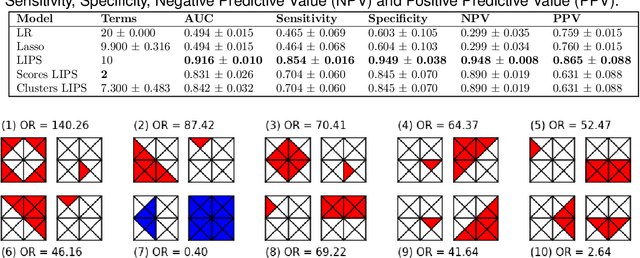
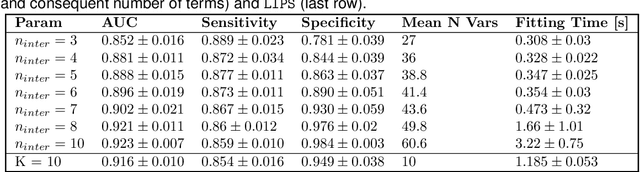
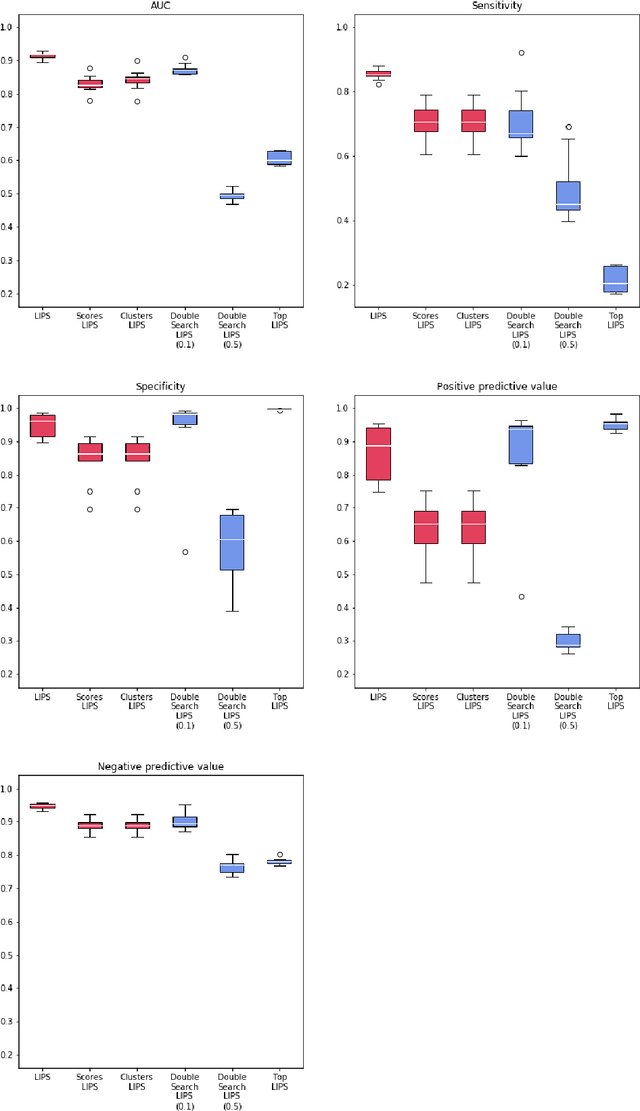
Abstract:Logistic Regression (LR) is a widely used statistical method in empirical binary classification studies. However, real-life scenarios oftentimes share complexities that prevent from the use of the as-is LR model, and instead highlight the need to include high-order interactions to capture data variability. This becomes even more challenging because of: (i) datasets growing wider, with more and more variables; (ii) studies being typically conducted in strongly imbalanced settings; (iii) samples going from very large to extremely small; (iv) the need of providing both predictive models and interpretable results. In this paper we present a novel algorithm, Learning high-order Interactions via targeted Pattern Search (LIPS), to select interaction terms of varying order to include in a LR model for an imbalanced binary classification task when input data are categorical. LIPS's rationale stems from the duality between item sets and categorical interactions. The algorithm relies on an interaction learning step based on a well-known frequent item set mining algorithm, and a novel dissimilarity-based interaction selection step that allows the user to specify the number of interactions to be included in the LR model. In addition, we particularize two variants (Scores LIPS and Clusters LIPS), that can address even more specific needs. Through a set of experiments we validate our algorithm and prove its wide applicability to real-life research scenarios, showing that it outperforms a benchmark state-of-the-art algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge