Emilio Dorigatti
How Inverse Conditional Flows Can Serve as a Substitute for Distributional Regression
May 08, 2024



Abstract:Neural network representations of simple models, such as linear regression, are being studied increasingly to better understand the underlying principles of deep learning algorithms. However, neural representations of distributional regression models, such as the Cox model, have received little attention so far. We close this gap by proposing a framework for distributional regression using inverse flow transformations (DRIFT), which includes neural representations of the aforementioned models. We empirically demonstrate that the neural representations of models in DRIFT can serve as a substitute for their classical statistical counterparts in several applications involving continuous, ordered, time-series, and survival outcomes. We confirm that models in DRIFT empirically match the performance of several statistical methods in terms of estimation of partial effects, prediction, and aleatoric uncertainty quantification. DRIFT covers both interpretable statistical models and flexible neural networks opening up new avenues in both statistical modeling and deep learning.
Approximate Bayes Optimal Pseudo-Label Selection
Feb 20, 2023Abstract:Semi-supervised learning by self-training heavily relies on pseudo-label selection (PLS). The selection often depends on the initial model fit on labeled data. Early overfitting might thus be propagated to the final model by selecting instances with overconfident but erroneous predictions, often referred to as confirmation bias. This paper introduces BPLS, a Bayesian framework for PLS that aims to mitigate this issue. At its core lies a criterion for selecting instances to label: an analytical approximation of the posterior predictive of pseudo-samples. We derive this selection criterion by proving Bayes optimality of the posterior predictive of pseudo-samples. We further overcome computational hurdles by approximating the criterion analytically. Its relation to the marginal likelihood allows us to come up with an approximation based on Laplace's method and the Gaussian integral. We empirically assess BPLS for parametric generalized linear and non-parametric generalized additive models on simulated and real-world data. When faced with high-dimensional data prone to overfitting, BPLS outperforms traditional PLS methods.
Joint Debiased Representation and Image Clustering Learning with Self-Supervision
Sep 14, 2022



Abstract:Contrastive learning is among the most successful methods for visual representation learning, and its performance can be further improved by jointly performing clustering on the learned representations. However, existing methods for joint clustering and contrastive learning do not perform well on long-tailed data distributions, as majority classes overwhelm and distort the loss of minority classes, thus preventing meaningful representations to be learned. Motivated by this, we develop a novel joint clustering and contrastive learning framework by adapting the debiased contrastive loss to avoid under-clustering minority classes of imbalanced datasets. We show that our proposed modified debiased contrastive loss and divergence clustering loss improves the performance across multiple datasets and learning tasks. The source code is available at https://anonymous.4open.science/r/SSL-debiased-clustering
Improved proteasomal cleavage prediction with positive-unlabeled learning
Sep 14, 2022

Abstract:Accurate in silico modeling of the antigen processing pathway is crucial to enable personalized epitope vaccine design for cancer. An important step of such pathway is the degradation of the vaccine into smaller peptides by the proteasome, some of which are going to be presented to T cells by the MHC complex. While predicting MHC-peptide presentation has received a lot of attention recently, proteasomal cleavage prediction remains a relatively unexplored area in light of recent advances in high-throughput mass spectrometry-based MHC ligandomics. Moreover, as such experimental techniques do not allow to identify regions that cannot be cleaved, the latest predictors generate synthetic negative samples and treat them as true negatives when training, even though some of them could actually be positives. In this work, we thus present a new predictor trained with an expanded dataset and the solid theoretical underpinning of positive-unlabeled learning, achieving a new state-of-the-art in proteasomal cleavage prediction. The improved predictive capabilities will in turn enable more precise vaccine development improving the efficacy of epitope-based vaccines. Code and pretrained models are available at https://github.com/SchubertLab/proteasomal-cleavage-puupl.
Robust and Efficient Imbalanced Positive-Unlabeled Learning with Self-supervision
Sep 06, 2022



Abstract:Learning from positive and unlabeled (PU) data is a setting where the learner only has access to positive and unlabeled samples while having no information on negative examples. Such PU setting is of great importance in various tasks such as medical diagnosis, social network analysis, financial markets analysis, and knowledge base completion, which also tend to be intrinsically imbalanced, i.e., where most examples are actually negatives. Most existing approaches for PU learning, however, only consider artificially balanced datasets and it is unclear how well they perform in the realistic scenario of imbalanced and long-tail data distribution. This paper proposes to tackle this challenge via robust and efficient self-supervised pretraining. However, training conventional self-supervised learning methods when applied with highly imbalanced PU distribution needs better reformulation. In this paper, we present \textit{ImPULSeS}, a unified representation learning framework for \underline{Im}balanced \underline{P}ositive \underline{U}nlabeled \underline{L}earning leveraging \underline{Se}lf-\underline{S}upervised debiase pre-training. ImPULSeS uses a generic combination of large-scale unsupervised learning with debiased contrastive loss and additional reweighted PU loss. We performed different experiments across multiple datasets to show that ImPULSeS is able to halve the error rate of the previous state-of-the-art, even compared with previous methods that are given the true prior. Moreover, our method showed increased robustness to prior misspecification and superior performance even when pretraining was performed on an unrelated dataset. We anticipate such robustness and efficiency will make it much easier for practitioners to obtain excellent results on other PU datasets of interest. The source code is available at \url{https://github.com/JSchweisthal/ImPULSeS}
Positive-Unlabeled Learning with Uncertainty-aware Pseudo-label Selection
Jan 31, 2022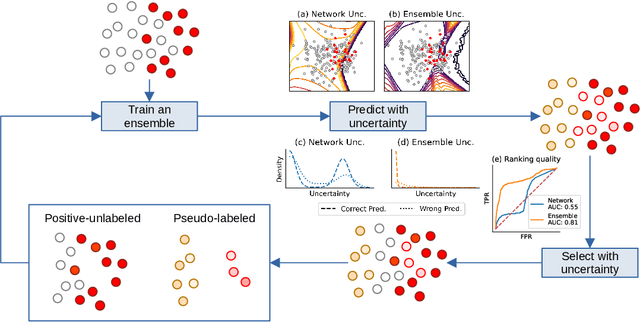
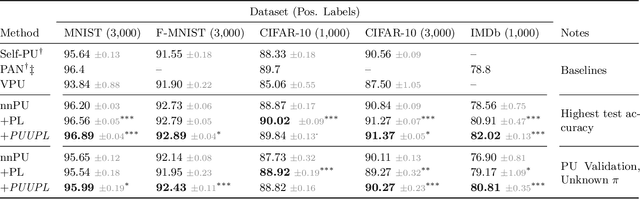
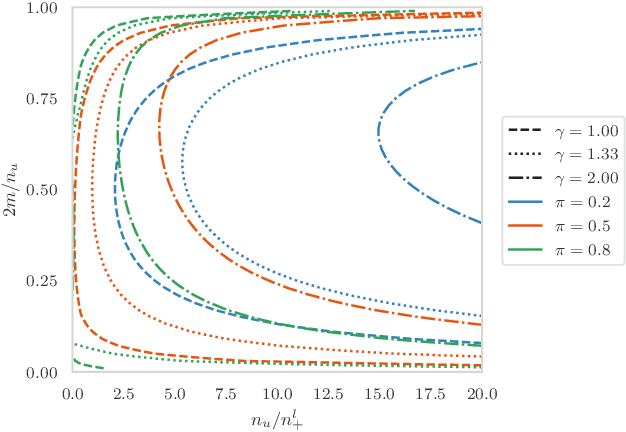
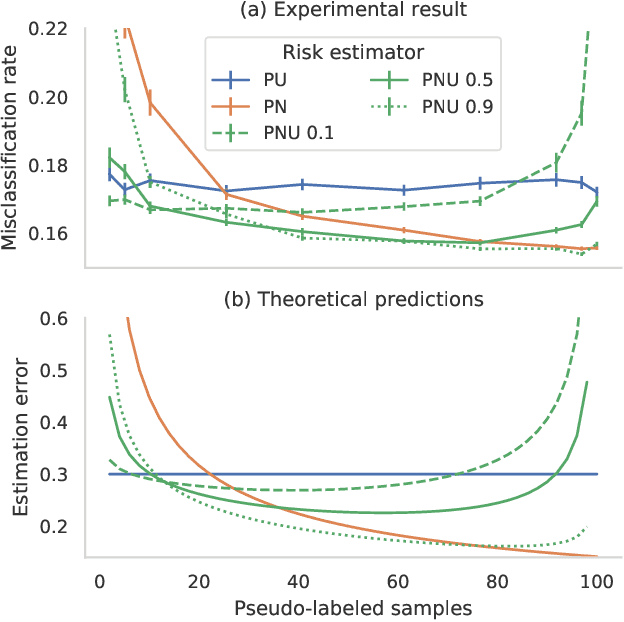
Abstract:Pseudo-labeling solutions for positive-unlabeled (PU) learning have the potential to result in higher performance compared to cost-sensitive learning but are vulnerable to incorrectly estimated pseudo-labels. In this paper, we provide a theoretical analysis of a risk estimator that combines risk on PU and pseudo-labeled data. Furthermore, we show analytically as well as experimentally that such an estimator results in lower excess risk compared to using PU data alone, provided that enough samples are pseudo-labeled with acceptable error rates. We then propose PUUPL, a novel training procedure for PU learning that leverages the epistemic uncertainty of an ensemble of deep neural networks to minimize errors in pseudo-label selection. We conclude with extensive experiments showing the effectiveness of our proposed algorithm over different datasets, modalities, and learning tasks. These show that PUUPL enables a reduction of up to 20% in test error rates even when prior and negative samples are not provided for validation, setting a new state-of-the-art for PU learning.
Learning Statistical Representation with Joint Deep Embedded Clustering
Sep 11, 2021



Abstract:One of the most promising approaches for unsupervised learning is combining deep representation learning and deep clustering. Some recent works propose to simultaneously learn representation using deep neural networks and perform clustering by defining a clustering loss on top of embedded features. However, these approaches are sensitive to imbalanced data and out-of-distribution samples. Hence, these methods optimize clustering by pushing data close to randomly initialized cluster centers. This is problematic when the number of instances varies largely in different classes or a cluster with few samples has less chance to be assigned a good centroid. To overcome these limitations, we introduce StatDEC, a new unsupervised framework for joint statistical representation learning and clustering. StatDEC simultaneously trains two deep learning models, a deep statistics network that captures the data distribution, and a deep clustering network that learns embedded features and performs clustering by explicitly defining a clustering loss. Specifically, the clustering network and representation network both take advantage of our proposed statistics pooling layer that represents mean, variance, and cardinality to handle the out-of-distribution samples as well as a class imbalance. Our experiments show that using these representations, one can considerably improve results on imbalanced image clustering across a variety of image datasets. Moreover, the learned representations generalize well when transferred to the out-of-distribution dataset.
Combining Graph Neural Networks and Spatio-temporal Disease Models to Predict COVID-19 Cases in Germany
Jan 03, 2021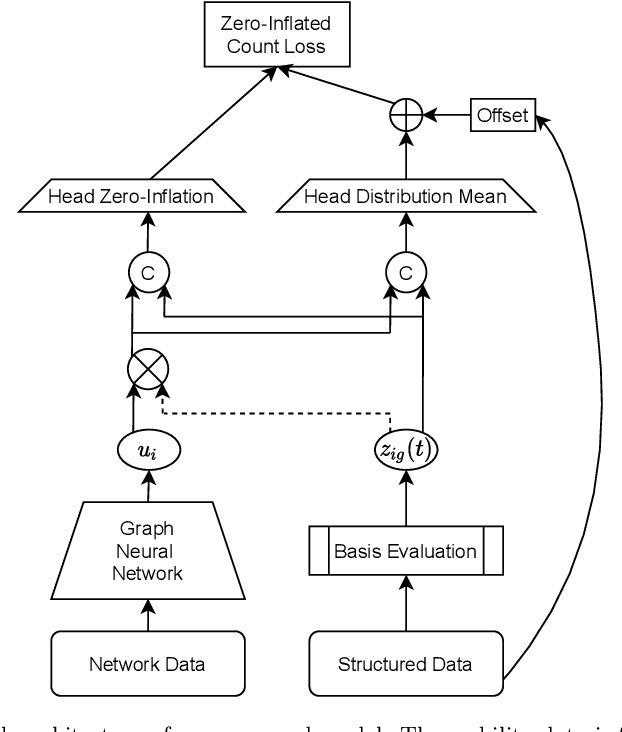
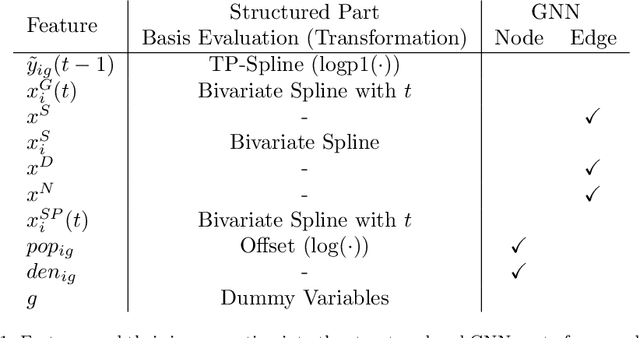
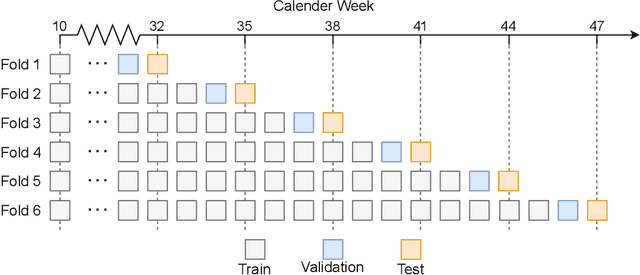
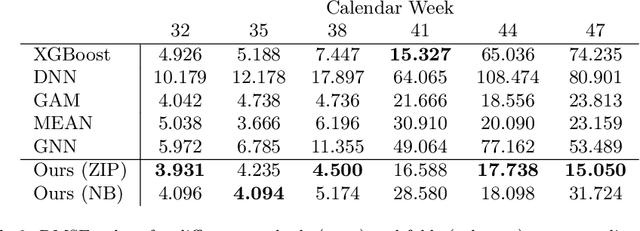
Abstract:During 2020, the infection rate of COVID-19 has been investigated by many scholars from different research fields. In this context, reliable and interpretable forecasts of disease incidents are a vital tool for policymakers to manage healthcare resources. Several experts have called for the necessity to account for human mobility to explain the spread of COVID-19. Existing approaches are often applying standard models of the respective research field. This habit, however, often comes along with certain restrictions. For instance, most statistical or epidemiological models cannot directly incorporate unstructured data sources, including relational data that may encode human mobility. In contrast, machine learning approaches may yield better predictions by exploiting these data structures, yet lack intuitive interpretability as they are often categorized as black-box models. We propose a trade-off between both research directions and present a multimodal learning approach that combines the advantages of statistical regression and machine learning models for predicting local COVID-19 cases in Germany. This novel approach enables the use of a richer collection of data types, including mobility flows and colocation probabilities, and yields the lowest MSE scores throughout our observational period in our benchmark study. The results corroborate the necessity of including mobility data and showcase the flexibility and interpretability of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge