Benjamin Schubert
Improved proteasomal cleavage prediction with positive-unlabeled learning
Sep 14, 2022

Abstract:Accurate in silico modeling of the antigen processing pathway is crucial to enable personalized epitope vaccine design for cancer. An important step of such pathway is the degradation of the vaccine into smaller peptides by the proteasome, some of which are going to be presented to T cells by the MHC complex. While predicting MHC-peptide presentation has received a lot of attention recently, proteasomal cleavage prediction remains a relatively unexplored area in light of recent advances in high-throughput mass spectrometry-based MHC ligandomics. Moreover, as such experimental techniques do not allow to identify regions that cannot be cleaved, the latest predictors generate synthetic negative samples and treat them as true negatives when training, even though some of them could actually be positives. In this work, we thus present a new predictor trained with an expanded dataset and the solid theoretical underpinning of positive-unlabeled learning, achieving a new state-of-the-art in proteasomal cleavage prediction. The improved predictive capabilities will in turn enable more precise vaccine development improving the efficacy of epitope-based vaccines. Code and pretrained models are available at https://github.com/SchubertLab/proteasomal-cleavage-puupl.
Positive-Unlabeled Learning with Uncertainty-aware Pseudo-label Selection
Jan 31, 2022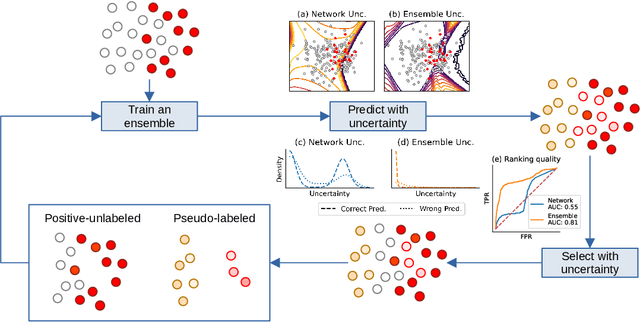
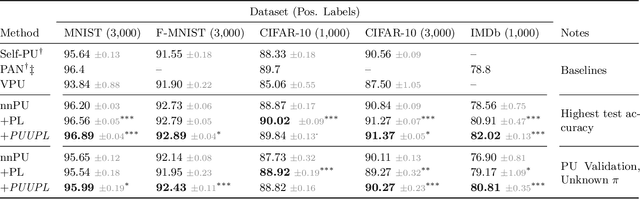
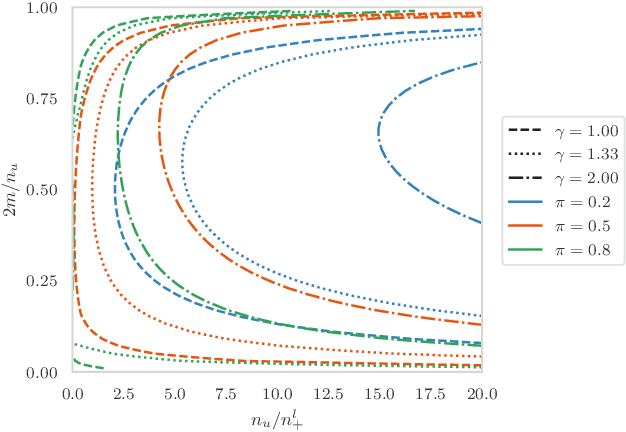
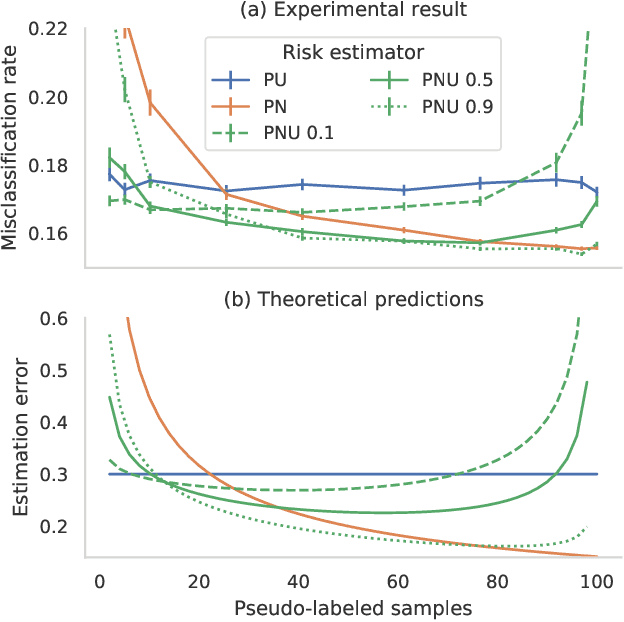
Abstract:Pseudo-labeling solutions for positive-unlabeled (PU) learning have the potential to result in higher performance compared to cost-sensitive learning but are vulnerable to incorrectly estimated pseudo-labels. In this paper, we provide a theoretical analysis of a risk estimator that combines risk on PU and pseudo-labeled data. Furthermore, we show analytically as well as experimentally that such an estimator results in lower excess risk compared to using PU data alone, provided that enough samples are pseudo-labeled with acceptable error rates. We then propose PUUPL, a novel training procedure for PU learning that leverages the epistemic uncertainty of an ensemble of deep neural networks to minimize errors in pseudo-label selection. We conclude with extensive experiments showing the effectiveness of our proposed algorithm over different datasets, modalities, and learning tasks. These show that PUUPL enables a reduction of up to 20% in test error rates even when prior and negative samples are not provided for validation, setting a new state-of-the-art for PU learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge