Elisabeth Gassiat
LMO
Membership Inference Risks in Quantized Models: A Theoretical and Empirical Study
Feb 10, 2025



Abstract:Quantizing machine learning models has demonstrated its effectiveness in lowering memory and inference costs while maintaining performance levels comparable to the original models. In this work, we investigate the impact of quantization procedures on the privacy of data-driven models, specifically focusing on their vulnerability to membership inference attacks. We derive an asymptotic theoretical analysis of Membership Inference Security (MIS), characterizing the privacy implications of quantized algorithm weights against the most powerful (and possibly unknown) attacks. Building on these theoretical insights, we propose a novel methodology to empirically assess and rank the privacy levels of various quantization procedures. Using synthetic datasets, we demonstrate the effectiveness of our approach in assessing the MIS of different quantizers. Furthermore, we explore the trade-off between privacy and performance using real-world data and models in the context of molecular modeling.
Tree-based variational inference for Poisson log-normal models
Jun 25, 2024Abstract:When studying ecosystems, hierarchical trees are often used to organize entities based on proximity criteria, such as the taxonomy in microbiology, social classes in geography, or product types in retail businesses, offering valuable insights into entity relationships. Despite their significance, current count-data models do not leverage this structured information. In particular, the widely used Poisson log-normal (PLN) model, known for its ability to model interactions between entities from count data, lacks the possibility to incorporate such hierarchical tree structures, limiting its applicability in domains characterized by such complexities. To address this matter, we introduce the PLN-Tree model as an extension of the PLN model, specifically designed for modeling hierarchical count data. By integrating structured variational inference techniques, we propose an adapted training procedure and establish identifiability results, enhancisng both theoretical foundations and practical interpretability. Additionally, we extend our framework to classification tasks as a preprocessing pipeline, showcasing its versatility. Experimental evaluations on synthetic datasets as well as real-world microbiome data demonstrate the superior performance of the PLN-Tree model in capturing hierarchical dependencies and providing valuable insights into complex data structures, showing the practical interest of knowledge graphs like the taxonomy in ecosystems modeling.
Fundamental Limits of Membership Inference Attacks on Machine Learning Models
Oct 27, 2023Abstract:Membership inference attacks (MIA) can reveal whether a particular data point was part of the training dataset, potentially exposing sensitive information about individuals. This article explores the fundamental statistical limitations associated with MIAs on machine learning models. More precisely, we first derive the statistical quantity that governs the effectiveness and success of such attacks. Then, we investigate several situations for which we provide bounds on this quantity of interest. This allows us to infer the accuracy of potential attacks as a function of the number of samples and other structural parameters of learning models, which in some cases can be directly estimated from the dataset.
Model-based clustering using non-parametric Hidden Markov Models
Sep 25, 2023Abstract:Thanks to their dependency structure, non-parametric Hidden Markov Models (HMMs) are able to handle model-based clustering without specifying group distributions. The aim of this work is to study the Bayes risk of clustering when using HMMs and to propose associated clustering procedures. We first give a result linking the Bayes risk of classification and the Bayes risk of clustering, which we use to identify the key quantity determining the difficulty of the clustering task. We also give a proof of this result in the i.i.d. framework, which might be of independent interest. Then we study the excess risk of the plugin classifier. All these results are shown to remain valid in the online setting where observations are clustered sequentially. Simulations illustrate our findings.
Fundamental limits for learning hidden Markov model parameters
Jun 24, 2021Abstract:We study the frontier between learnable and unlearnable hidden Markov models (HMMs). HMMs are flexible tools for clustering dependent data coming from unknown populations. The model parameters are known to be identifiable as soon as the clusters are distinct and the hidden chain is ergodic with a full rank transition matrix. In the limit as any one of these conditions fails, it becomes impossible to identify parameters. For a chain with two hidden states we prove nonasymptotic minimax upper and lower bounds, matching up to constants, which exhibit thresholds at which the parameters become learnable.
Disentangling Identifiable Features from Noisy Data with Structured Nonlinear ICA
Jun 17, 2021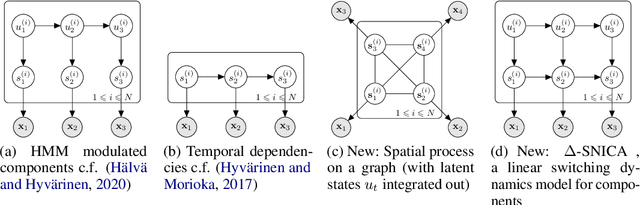
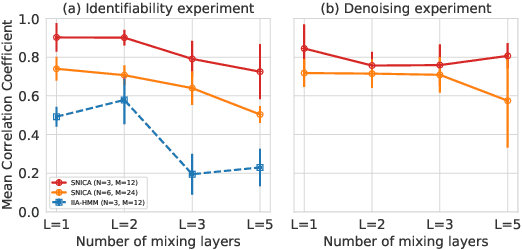

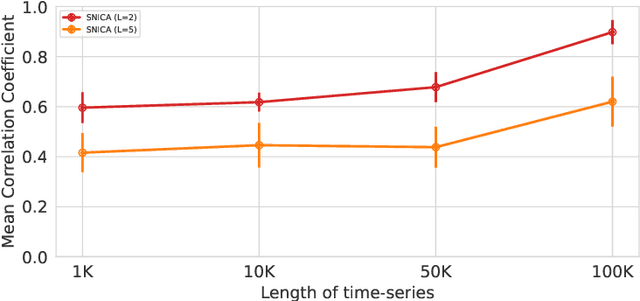
Abstract:We introduce a new general identifiable framework for principled disentanglement referred to as Structured Nonlinear Independent Component Analysis (SNICA). Our contribution is to extend the identifiability theory of deep generative models for a very broad class of structured models. While previous works have shown identifiability for specific classes of time-series models, our theorems extend this to more general temporal structures as well as to models with more complex structures such as spatial dependencies. In particular, we establish the major result that identifiability for this framework holds even in the presence of noise of unknown distribution. The SNICA setting therefore subsumes all the existing nonlinear ICA models for time-series and also allows for new much richer identifiable models. Finally, as an example of our framework's flexibility, we introduce the first nonlinear ICA model for time-series that combines the following very useful properties: it accounts for both nonstationarity and autocorrelation in a fully unsupervised setting; performs dimensionality reduction; models hidden states; and enables principled estimation and inference by variational maximum-likelihood.
Joint self-supervised blind denoising and noise estimation
Feb 16, 2021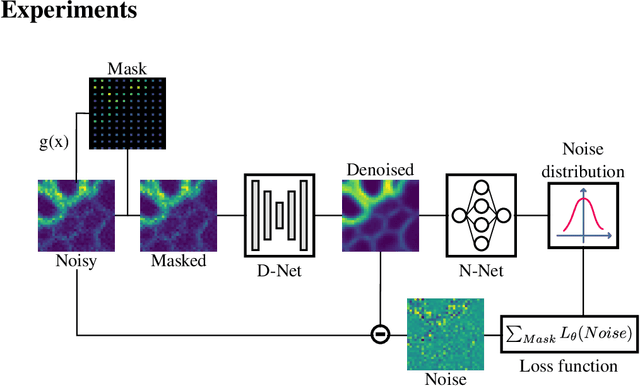



Abstract:We propose a novel self-supervised image blind denoising approach in which two neural networks jointly predict the clean signal and infer the noise distribution. Assuming that the noisy observations are independent conditionally to the signal, the networks can be jointly trained without clean training data. Therefore, our approach is particularly relevant for biomedical image denoising where the noise is difficult to model precisely and clean training data are usually unavailable. Our method significantly outperforms current state-of-the-art self-supervised blind denoising algorithms, on six publicly available biomedical image datasets. We also show empirically with synthetic noisy data that our model captures the noise distribution efficiently. Finally, the described framework is simple, lightweight and computationally efficient, making it useful in practical cases.
Multinomial logistic model for coinfection diagnosis between arbovirus and malaria in Kedougou
Jan 12, 2018



Abstract:In tropical regions, populations continue to suffer morbidity and mortality from malaria and arboviral diseases. In Kedougou (Senegal), these illnesses are all endemic due to the climate and its geographical position. The co-circulation of malaria parasites and arboviruses can explain the observation of coinfected cases. Indeed there is strong resemblance in symptoms between these diseases making problematic targeted medical care of coinfected cases. This is due to the fact that the origin of illness is not obviously known. Some cases could be immunized against one or the other of the pathogens, immunity typically acquired with factors like age and exposure as usual for endemic area. Then, coinfection needs to be better diagnosed. Using data collected from patients in Kedougou region, from 2009 to 2013, we adjusted a multinomial logistic model and selected relevant variables in explaining coinfection status. We observed specific sets of variables explaining each of the diseases exclusively and the coinfection. We tested the independence between arboviral and malaria infections and derived coinfection probabilities from the model fitting. In case of a coinfection probability greater than a threshold value to be calibrated on the data, duration of illness above 3 days and age above 10 years-old are mostly indicative of arboviral disease while body temperature higher than 40{\textdegree}C and presence of nausea or vomiting symptoms during the rainy season are mostly indicative of malaria disease.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge