Jean Ollion
CMAP
DistNet2D: Leveraging long-range temporal information for efficient segmentation and tracking
Oct 30, 2023Abstract:Extracting long tracks and lineages from videomicroscopy requires an extremely low error rate, which is challenging on complex datasets of dense or deforming cells. Leveraging temporal context is key to overcome this challenge. We propose DistNet2D, a new deep neural network (DNN) architecture for 2D cell segmentation and tracking that leverages both mid- and long-term temporal context. DistNet2D considers seven frames at the input and uses a post-processing procedure that exploits information from the entire movie to correct segmentation errors. DistNet2D outperforms two recent methods on two experimental datasets, one containing densely packed bacterial cells and the other containing eukaryotic cells. It has been integrated into an ImageJ-based graphical user interface for 2D data visualization, curation, and training. Finally, we demonstrate the performance of DistNet2D on correlating the size and shape of cells with their transport properties over large statistics, for both bacterial and eukaryotic cells.
Joint self-supervised blind denoising and noise estimation
Feb 16, 2021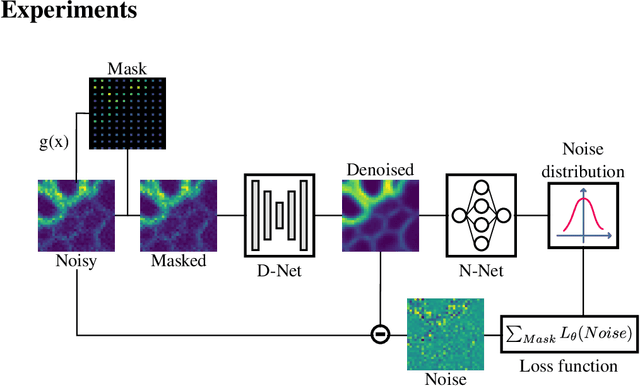



Abstract:We propose a novel self-supervised image blind denoising approach in which two neural networks jointly predict the clean signal and infer the noise distribution. Assuming that the noisy observations are independent conditionally to the signal, the networks can be jointly trained without clean training data. Therefore, our approach is particularly relevant for biomedical image denoising where the noise is difficult to model precisely and clean training data are usually unavailable. Our method significantly outperforms current state-of-the-art self-supervised blind denoising algorithms, on six publicly available biomedical image datasets. We also show empirically with synthetic noisy data that our model captures the noise distribution efficiently. Finally, the described framework is simple, lightweight and computationally efficient, making it useful in practical cases.
DistNet: Deep Tracking by displacement regression: application to bacteria growing in the Mother Machine
Mar 17, 2020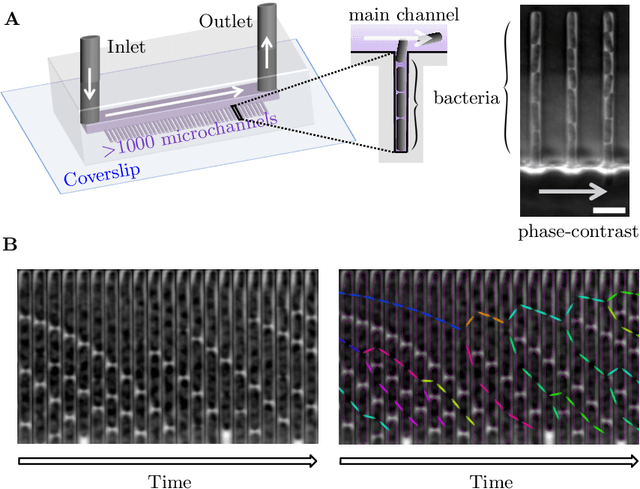
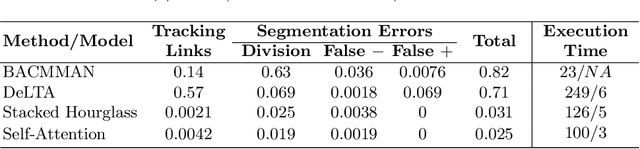
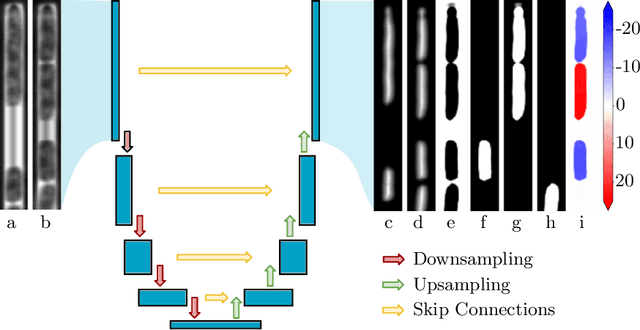
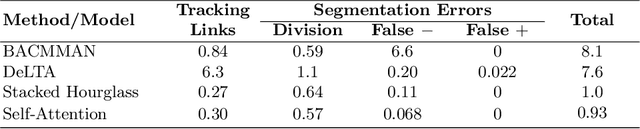
Abstract:The mother machine is a popular microfluidic device that allows long-term time-lapse imaging of thousands of cells in parallel by microscopy. It has become a valuable tool for single-cell level quantitative analysis and characterization of many cellular processes such as gene expression and regulation, mutagenesis or response to antibiotics. The automated and quantitative analysis of the massive amount of data generated by such experiments is now the limiting step. In particular the segmentation and tracking of bacteria cells imaged in phase-contrast microscopy---with error rates compatible with high-throughput data---is a challenging problem. In this work, we describe a novel formulation of the multi-object tracking problem, in which tracking is performed by a regression of the bacteria's displacement, allowing simultaneous tracking of multiple bacteria, despite their growth and division over time. Our method performs jointly segmentation and tracking, leveraging sequential information to increase segmentation accuracy. We introduce a Deep Neural Network architecture taking advantage of a self-attention mechanism which yields less than 0.005% tracking error rate and less than 0.03% segmentation error rate. We demonstrate superior performance and speed compared to state-of-the-art methods. While this method is particularly well suited for mother machine microscopy data, its general joint tracking and segmentation formulation could be applied to many other problems with different geometries.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge