Elaine Angelino
Learning Certifiably Optimal Rule Lists for Categorical Data
Aug 03, 2018



Abstract:We present the design and implementation of a custom discrete optimization technique for building rule lists over a categorical feature space. Our algorithm produces rule lists with optimal training performance, according to the regularized empirical risk, with a certificate of optimality. By leveraging algorithmic bounds, efficient data structures, and computational reuse, we achieve several orders of magnitude speedup in time and a massive reduction of memory consumption. We demonstrate that our approach produces optimal rule lists on practical problems in seconds. Our results indicate that it is possible to construct optimal sparse rule lists that are approximately as accurate as the COMPAS proprietary risk prediction tool on data from Broward County, Florida, but that are completely interpretable. This framework is a novel alternative to CART and other decision tree methods for interpretable modeling.
* A short version of this work appeared in KDD '17 as "Learning Certifiably Optimal Rule Lists"
Patterns of Scalable Bayesian Inference
Mar 22, 2016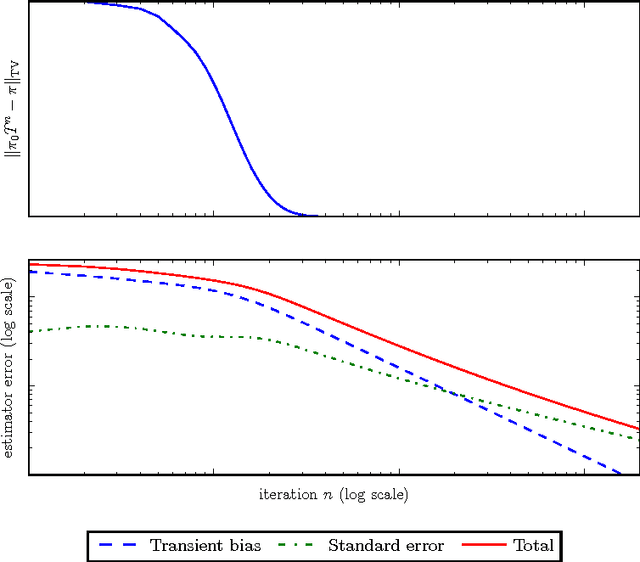

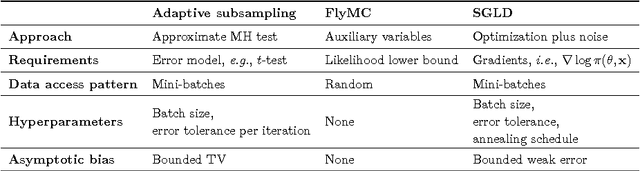
Abstract:Datasets are growing not just in size but in complexity, creating a demand for rich models and quantification of uncertainty. Bayesian methods are an excellent fit for this demand, but scaling Bayesian inference is a challenge. In response to this challenge, there has been considerable recent work based on varying assumptions about model structure, underlying computational resources, and the importance of asymptotic correctness. As a result, there is a zoo of ideas with few clear overarching principles. In this paper, we seek to identify unifying principles, patterns, and intuitions for scaling Bayesian inference. We review existing work on utilizing modern computing resources with both MCMC and variational approximation techniques. From this taxonomy of ideas, we characterize the general principles that have proven successful for designing scalable inference procedures and comment on the path forward.
Variational consensus Monte Carlo
Jun 09, 2015


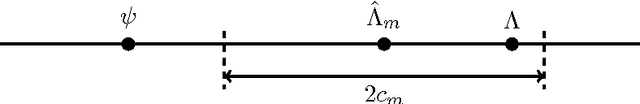
Abstract:Practitioners of Bayesian statistics have long depended on Markov chain Monte Carlo (MCMC) to obtain samples from intractable posterior distributions. Unfortunately, MCMC algorithms are typically serial, and do not scale to the large datasets typical of modern machine learning. The recently proposed consensus Monte Carlo algorithm removes this limitation by partitioning the data and drawing samples conditional on each partition in parallel (Scott et al, 2013). A fixed aggregation function then combines these samples, yielding approximate posterior samples. We introduce variational consensus Monte Carlo (VCMC), a variational Bayes algorithm that optimizes over aggregation functions to obtain samples from a distribution that better approximates the target. The resulting objective contains an intractable entropy term; we therefore derive a relaxation of the objective and show that the relaxed problem is blockwise concave under mild conditions. We illustrate the advantages of our algorithm on three inference tasks from the literature, demonstrating both the superior quality of the posterior approximation and the moderate overhead of the optimization step. Our algorithm achieves a relative error reduction (measured against serial MCMC) of up to 39% compared to consensus Monte Carlo on the task of estimating 300-dimensional probit regression parameter expectations; similarly, it achieves an error reduction of 92% on the task of estimating cluster comembership probabilities in a Gaussian mixture model with 8 components in 8 dimensions. Furthermore, these gains come at moderate cost compared to the runtime of serial MCMC, achieving near-ideal speedup in some instances.
Attribute-Efficient Evolvability of Linear Functions
Apr 03, 2014
Abstract:In a seminal paper, Valiant (2006) introduced a computational model for evolution to address the question of complexity that can arise through Darwinian mechanisms. Valiant views evolution as a restricted form of computational learning, where the goal is to evolve a hypothesis that is close to the ideal function. Feldman (2008) showed that (correlational) statistical query learning algorithms could be framed as evolutionary mechanisms in Valiant's model. P. Valiant (2012) considered evolvability of real-valued functions and also showed that weak-optimization algorithms that use weak-evaluation oracles could be converted to evolutionary mechanisms. In this work, we focus on the complexity of representations of evolutionary mechanisms. In general, the reductions of Feldman and P. Valiant may result in intermediate representations that are arbitrarily complex (polynomial-sized circuits). We argue that biological constraints often dictate that the representations have low complexity, such as constant depth and fan-in circuits. We give mechanisms for evolving sparse linear functions under a large class of smooth distributions. These evolutionary algorithms are attribute-efficient in the sense that the size of the representations and the number of generations required depend only on the sparsity of the target function and the accuracy parameter, but have no dependence on the total number of attributes.
Accelerating MCMC via Parallel Predictive Prefetching
Mar 28, 2014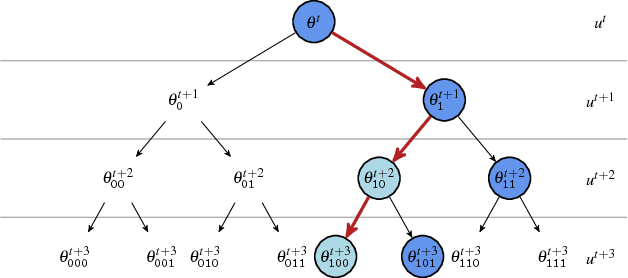
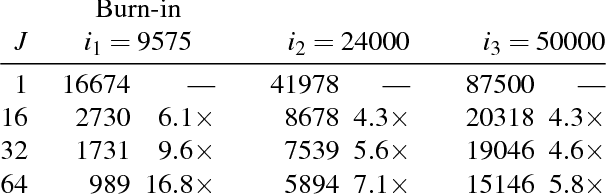


Abstract:We present a general framework for accelerating a large class of widely used Markov chain Monte Carlo (MCMC) algorithms. Our approach exploits fast, iterative approximations to the target density to speculatively evaluate many potential future steps of the chain in parallel. The approach can accelerate computation of the target distribution of a Bayesian inference problem, without compromising exactness, by exploiting subsets of data. It takes advantage of whatever parallel resources are available, but produces results exactly equivalent to standard serial execution. In the initial burn-in phase of chain evaluation, it achieves speedup over serial evaluation that is close to linear in the number of available cores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge