Derek K. Jones
Shining light on degeneracies and uncertainties in quantifying both exchange and restriction with time-dependent diffusion MRI using Bayesian inference
Aug 26, 2025Abstract:Diffusion MRI (dMRI) biophysical models hold promise for characterizing gray matter tissue microstructure. Yet, the reliability of estimated parameters remains largely under-studied, especially in models that incorporate water exchange. In this study, we investigate the accuracy, precision, and presence of degeneracy of two recently proposed gray matter models, NEXI and SANDIX, using two acquisition protocols from the literature, on both simulated and in vivo data. We employ $\mu$GUIDE, a Bayesian inference framework based on deep learning, to quantify model uncertainty and detect parameter degeneracies, enabling a more interpretable assessment of fitted parameters. Our results show that while some microstructural parameters, such as extra-cellular diffusivity and neurite signal fraction, are robustly estimated, others, such as exchange time and soma radius, are often associated with high uncertainty and estimation bias, especially under realistic noise conditions and reduced acquisition protocols. Comparisons with non-linear least squares fitting underscore the added value of uncertainty-aware methods, which allow for the identification and filtering of unreliable estimates. These findings emphasize the need to report uncertainty and consider model degeneracies when interpreting model-based estimates. Our study advocates for the integration of probabilistic fitting approaches in neuroscience imaging pipelines to improve reproducibility and biological interpretability.
Image Quality Transfer of Diffusion MRI Guided By High-Resolution Structural MRI
Aug 06, 2024



Abstract:Prior work on the Image Quality Transfer on Diffusion MRI (dMRI) has shown significant improvement over traditional interpolation methods. However, the difficulty in obtaining ultra-high resolution Diffusion MRI scans poses a problem in training neural networks to obtain high-resolution dMRI scans. Here we hypothesise that the inclusion of structural MRI images, which can be acquired at much higher resolutions, can be used as a guide to obtaining a more accurate high-resolution dMRI output. To test our hypothesis, we have constructed a novel framework that incorporates structural MRI scans together with dMRI to obtain high-resolution dMRI scans. We set up tests which evaluate the validity of our claim through various configurations and compare the performance of our approach against a unimodal approach. Our results show that the inclusion of structural MRI scans do lead to an improvement in high-resolution image prediction when T1w data is incorporated into the model input.
Lossy compression of multidimensional medical images using sinusoidal activation networks: an evaluation study
Aug 03, 2022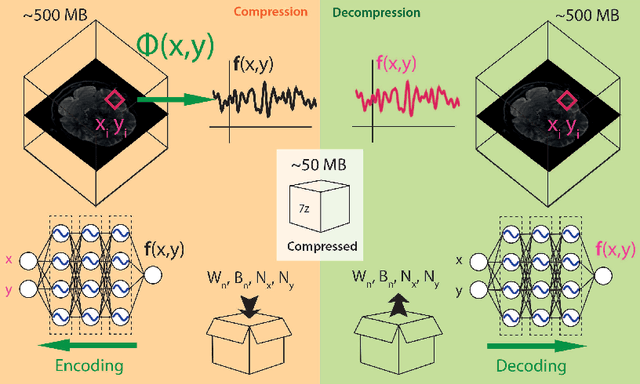

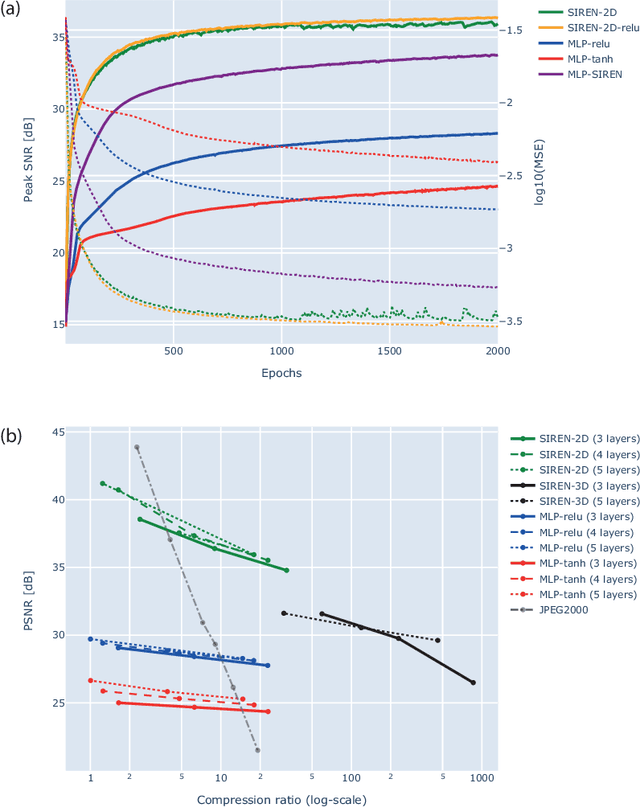
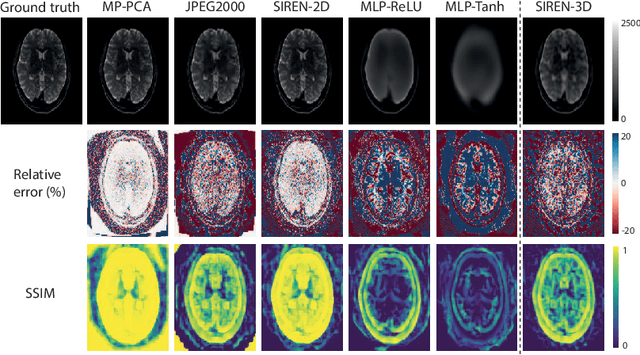
Abstract:In this work, we evaluate how neural networks with periodic activation functions can be leveraged to reliably compress large multidimensional medical image datasets, with proof-of-concept application to 4D diffusion-weighted MRI (dMRI). In the medical imaging landscape, multidimensional MRI is a key area of research for developing biomarkers that are both sensitive and specific to the underlying tissue microstructure. However, the high-dimensional nature of these data poses a challenge in terms of both storage and sharing capabilities and associated costs, requiring appropriate algorithms able to represent the information in a low-dimensional space. Recent theoretical developments in deep learning have shown how periodic activation functions are a powerful tool for implicit neural representation of images and can be used for compression of 2D images. Here we extend this approach to 4D images and show how any given 4D dMRI dataset can be accurately represented through the parameters of a sinusoidal activation network, achieving a data compression rate about 10 times higher than the standard DEFLATE algorithm. Our results show that the proposed approach outperforms benchmark ReLU and Tanh activation perceptron architectures in terms of mean squared error, peak signal-to-noise ratio and structural similarity index. Subsequent analyses using the tensor and spherical harmonics representations demonstrate that the proposed lossy compression reproduces accurately the characteristics of the original data, leading to relative errors about 5 to 10 times lower than the benchmark JPEG2000 lossy compression and similar to standard pre-processing steps such as MP-PCA denosing, suggesting a loss of information within the currently accepted levels for clinical application.
aDWI-BIDS: an extension to the brain imaging data structure for advanced diffusion weighted imaging
Apr 12, 2021

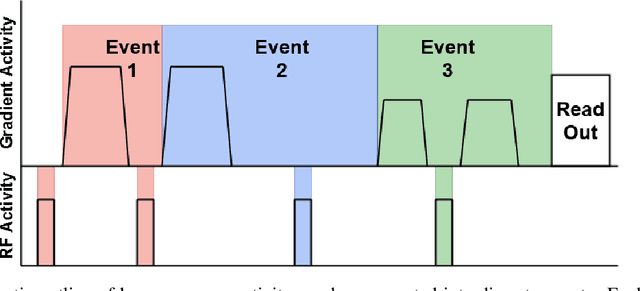
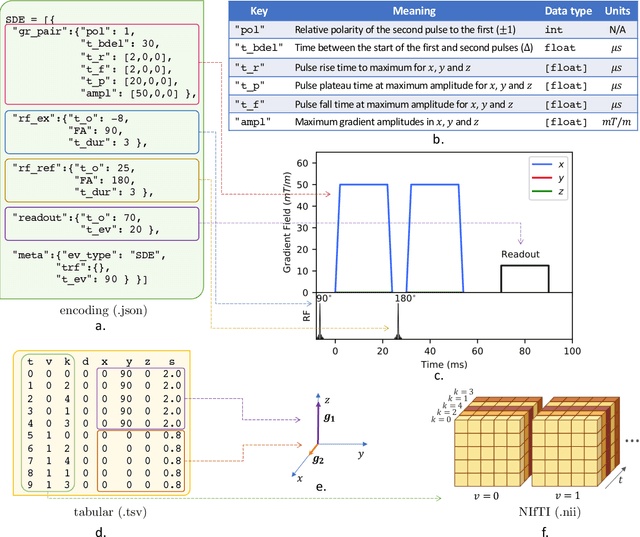
Abstract:Diffusion weighted imaging techniques permit us to infer microstructural detail in biological tissue in vivo and noninvasively. Modern sequences are based on advanced diffusion encoding schemes, allowing probing of more revealing measures of tissue microstructure than the standard apparent diffusion coefficient or fractional anisotropy. Though these methods may result in faster or more revealing acquisitions, they generally demand prior knowledge of sequence-specific parameters for which there is no accepted sharing standard. Here, we present a metadata labelling scheme suitable for the needs of developers and users within the diffusion neuroimaging community alike: a lightweight, unambiguous parametric map relaying acqusition parameters. This extensible scheme supports a wide spectrum of diffusion encoding methods, from single diffusion encoding to highly complex sequences involving arbitrary gradient waveforms. Built under the brain imaging data structure (BIDS), it allows storage of advanced diffusion MRI data comprehensively alongside any other neuroimaging information, facilitating processing pipelines and multimodal analyses. We illustrate the usefulness of this BIDS-extension with a range of example data, and discuss the extension's impact on pre- and post-processing software.
Tractometry-based Anomaly Detection for Single-subject White Matter Analysis
May 25, 2020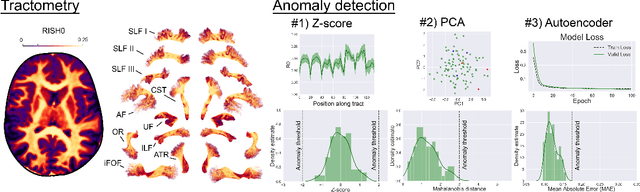
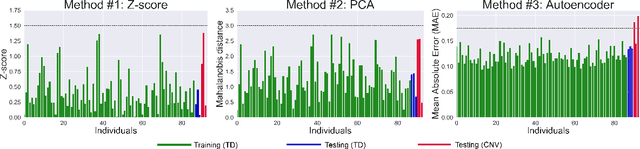
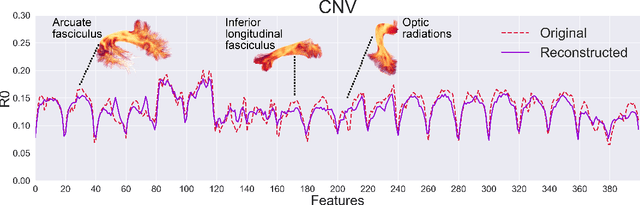
Abstract:There is an urgent need for a paradigm shift from group-wise comparisons to individual diagnosis in diffusion MRI (dMRI) to enable the analysis of rare cases and clinically-heterogeneous groups. Deep autoencoders have shown great potential to detect anomalies in neuroimaging data. We present a framework that operates on the manifold of white matter (WM) pathways to learn normative microstructural features, and discriminate those at genetic risk from controls in a paediatric population.
* Medical Imaging with Deep Learning (MIDL2020) Conference Short Paper
q-Space Novelty Detection with Variational Autoencoders
Oct 25, 2018


Abstract:In machine learning, novelty detection is the task of identifying novel unseen data. During training, only samples from the normal class are available. Test samples are classified as normal or abnormal by assignment of a novelty score. Here we propose novelty detection methods based on training variational autoencoders (VAEs) on normal data. Since abnormal samples are not used during training, we define novelty metrics based on the (partially complementary) assumptions that the VAE is less capable of reconstructing abnormal samples well; that abnormal samples more strongly violate the VAE regularizer; and that abnormal samples differ from normal samples not only in input-feature space, but also in the VAE latent space and VAE output. These approaches, combined with various possibilities of using (e.g. sampling) the probabilistic VAE to obtain scalar novelty scores, yield a large family of methods. We apply these methods to magnetic resonance imaging, namely to the detection of diffusion-space (q-space) abnormalities in diffusion MRI scans of multiple sclerosis patients, i.e. to detect multiple sclerosis lesions without using any lesion labels for training. Many of our methods outperform previously proposed q-space novelty detection methods. We also evaluate the proposed methods on the MNIST handwritten digits dataset and show that many of them are able to outperform the state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge