Filip Szczepankiewicz
MBD: Multi b-value Denoising of Diffusion Magnetic Resonance Images
Oct 22, 2024Abstract:We propose a novel approach to denoising diffusion magnetic resonance images (dMRI) using convolutional neural networks, that exploits the benefits of data acquired at multiple b-values to offset the need for many redundant observations. Denoising is especially relevant in dMRI since noise can have a deleterious impact on both quantification accuracy and image preprocessing. The most successful methods proposed to date, like Marchenko-Pastur Principal Component Analysis (MPPCA) denoising, are tailored to diffusion-weighting repeated for many encoding directions. They exploit high redundancy of the dataset that oversamples the diffusion-encoding direction space, since many directions have collinear components. However, there are many dMRI techniques that do not entail a large number of encoding directions or repetitions, and are therefore less suited to this approach. For example, clinical dMRI exams may include as few as three encoding directions, with low or negligible data redundancy across directions. Moreover, promising new dMRI approaches, like spherical b-tensor encoding (STE), benefit from high b-values while sensitizing the signal to diffusion along all directions in just a single shot. We introduce a convolutional neural network approach that we call multi-b-value-based denoising (MBD). MBD exploits the similarity in diffusion-weighted images (DWI) across different b-values but along the same diffusion encoding direction. It allows denoising of diffusion images with high noise variance while avoiding blurring, and using just a small number input images.
Geometry of the cumulant series in neuroimaging
Sep 04, 2024Abstract:Water diffusion gives rise to micrometer-scale sensitivity of diffusion MRI (dMRI) to cellular-level tissue structure. The advent of precision medicine and quantitative imaging hinges on revealing the information content of dMRI, and providing its parsimonious basis- and hardware-independent "fingerprint". Here we reveal the geometry of a multi-dimensional dMRI signal, classify all 21 invariants of diffusion and covariance tensors in terms of irreducible representations of the group of rotations, and relate them to tissue properties. Previously studied dMRI contrasts are expressed via 7 invariants, while the remaining 14 provide novel complementary information. We design acquisitions based on icosahedral vertices guaranteeing minimal number of measurements to determine 3-4 most used invariants in only 1-2 minutes for the whole brain. Representing dMRI signals via scalar invariant maps with definite symmetries will underpin machine learning classifiers of brain pathology, development, and aging, while fast protocols will enable translation of advanced dMRI into clinical practice.
Microstructural neuroimaging using spherical convolutional neural networks
Nov 17, 2022



Abstract:Diffusion-weighted magnetic resonance imaging is sensitive to the microstructural properties of brain tissue. However, estimating clinically and scientifically relevant microstructural properties from the measured signals remains a highly challenging inverse problem. This paper presents a novel framework for estimating microstructural parameters using recently developed orientationally invariant spherical convolutional neural networks and efficiently simulated training data with a known ground truth. The network was trained to predict the ground-truth parameter values from simulated noisy data and applied to imaging data acquired in a clinical setting to generate microstructural parameter maps. Our model could estimate model parameters from spherical data more accurately than conventional non-linear least squares or a multi-layer perceptron applied on powder-averaged data (i.e., the spherical mean technique, a popular method for orientationally invariant microstructural parameter estimation). Importantly, our method is generalizable and can be used to estimate the parameters of any Gaussian compartment model.
aDWI-BIDS: an extension to the brain imaging data structure for advanced diffusion weighted imaging
Apr 12, 2021

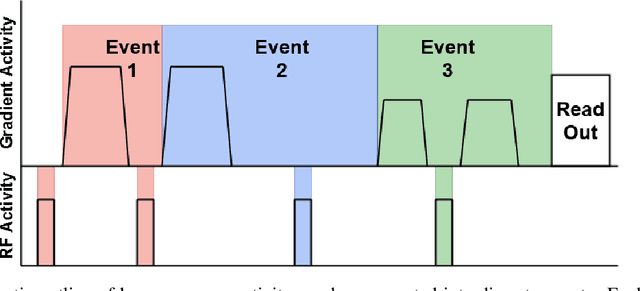
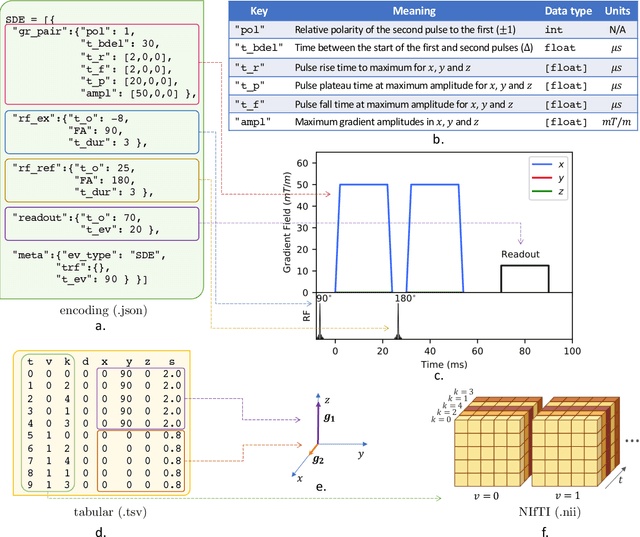
Abstract:Diffusion weighted imaging techniques permit us to infer microstructural detail in biological tissue in vivo and noninvasively. Modern sequences are based on advanced diffusion encoding schemes, allowing probing of more revealing measures of tissue microstructure than the standard apparent diffusion coefficient or fractional anisotropy. Though these methods may result in faster or more revealing acquisitions, they generally demand prior knowledge of sequence-specific parameters for which there is no accepted sharing standard. Here, we present a metadata labelling scheme suitable for the needs of developers and users within the diffusion neuroimaging community alike: a lightweight, unambiguous parametric map relaying acqusition parameters. This extensible scheme supports a wide spectrum of diffusion encoding methods, from single diffusion encoding to highly complex sequences involving arbitrary gradient waveforms. Built under the brain imaging data structure (BIDS), it allows storage of advanced diffusion MRI data comprehensively alongside any other neuroimaging information, facilitating processing pipelines and multimodal analyses. We illustrate the usefulness of this BIDS-extension with a range of example data, and discuss the extension's impact on pre- and post-processing software.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge