Davis J. McCarthy
Technical outlier detection via convolutional variational autoencoder for the ADMANI breast mammogram dataset
May 20, 2023
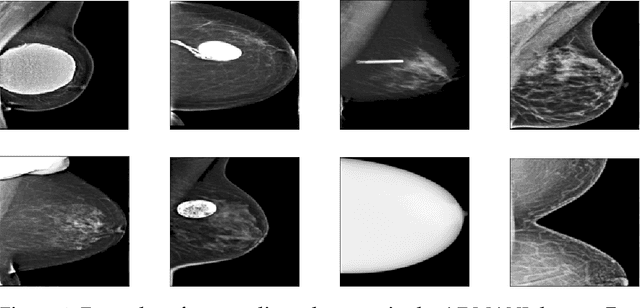

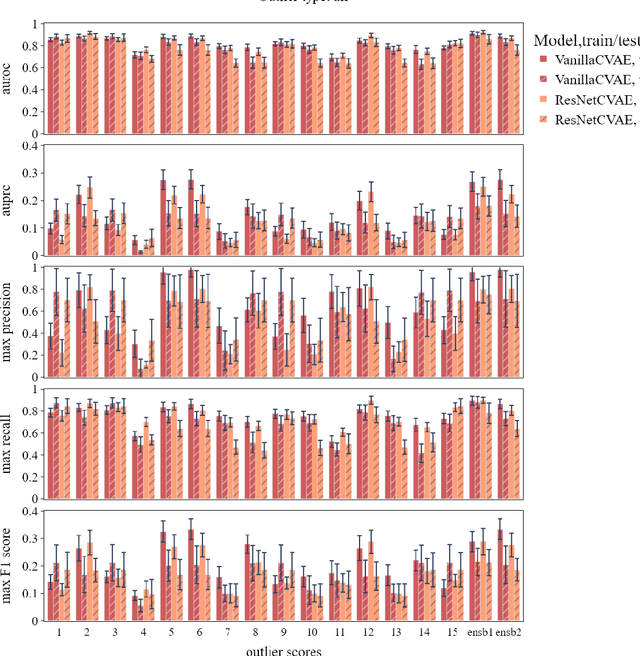
Abstract:The ADMANI datasets (annotated digital mammograms and associated non-image datasets) from the Transforming Breast Cancer Screening with AI programme (BRAIx) run by BreastScreen Victoria in Australia are multi-centre, large scale, clinically curated, real-world databases. The datasets are expected to aid in the development of clinically relevant Artificial Intelligence (AI) algorithms for breast cancer detection, early diagnosis, and other applications. To ensure high data quality, technical outliers must be removed before any downstream algorithm development. As a first step, we randomly select 30,000 individual mammograms and use Convolutional Variational Autoencoder (CVAE), a deep generative neural network, to detect outliers. CVAE is expected to detect all sorts of outliers, although its detection performance differs among different types of outliers. Traditional image processing techniques such as erosion and pectoral muscle analysis can compensate for the poor performance of CVAE in certain outlier types. We identify seven types of technical outliers: implant, pacemaker, cardiac loop recorder, improper radiography, atypical lesion/calcification, incorrect exposure parameter and improper placement. The outlier recall rate for the test set is 61% if CVAE, erosion and pectoral muscle analysis each select the top 1% images ranked in ascending or descending order according to image outlier score under each detection method, and 83% if each selects the top 5% images. This study offers an overview of technical outliers in the ADMANI dataset and suggests future directions to improve outlier detection effectiveness.
BRAIxDet: Learning to Detect Malignant Breast Lesion with Incomplete Annotations
Feb 02, 2023



Abstract:Methods to detect malignant lesions from screening mammograms are usually trained with fully annotated datasets, where images are labelled with the localisation and classification of cancerous lesions. However, real-world screening mammogram datasets commonly have a subset that is fully annotated and another subset that is weakly annotated with just the global classification (i.e., without lesion localisation). Given the large size of such datasets, researchers usually face a dilemma with the weakly annotated subset: to not use it or to fully annotate it. The first option will reduce detection accuracy because it does not use the whole dataset, and the second option is too expensive given that the annotation needs to be done by expert radiologists. In this paper, we propose a middle-ground solution for the dilemma, which is to formulate the training as a weakly- and semi-supervised learning problem that we refer to as malignant breast lesion detection with incomplete annotations. To address this problem, our new method comprises two stages, namely: 1) pre-training a multi-view mammogram classifier with weak supervision from the whole dataset, and 2) extending the trained classifier to become a multi-view detector that is trained with semi-supervised student-teacher learning, where the training set contains fully and weakly-annotated mammograms. We provide extensive detection results on two real-world screening mammogram datasets containing incomplete annotations, and show that our proposed approach achieves state-of-the-art results in the detection of malignant breast lesions with incomplete annotations.
Learning Support and Trivial Prototypes for Interpretable Image Classification
Jan 08, 2023



Abstract:Prototypical part network (ProtoPNet) methods have been designed to achieve interpretable classification by associating predictions with a set of training prototypes, which we refer to as trivial (i.e., easy-to-learn) prototypes because they are trained to lie far from the classification boundary in the feature space. Note that it is possible to make an analogy between ProtoPNet and support vector machine (SVM) given that the classification from both methods relies on computing similarity with a set of training points (i.e., trivial prototypes in ProtoPNet, and support vectors in SVM). However, while trivial prototypes are located far from the classification boundary, support vectors are located close to this boundary, and we argue that this discrepancy with the well-established SVM theory can result in ProtoPNet models with suboptimal classification accuracy. In this paper, we aim to improve the classification accuracy of ProtoPNet with a new method to learn support prototypes that lie near the classification boundary in the feature space, as suggested by the SVM theory. In addition, we target the improvement of classification interpretability with a new model, named ST-ProtoPNet, which exploits our support prototypes and the trivial prototypes to provide complementary interpretability information. Experimental results on CUB-200-2011, Stanford Cars, and Stanford Dogs datasets demonstrate that the proposed method achieves state-of-the-art classification accuracy and produces more visually meaningful and diverse prototypes.
Knowledge Distillation to Ensemble Global and Interpretable Prototype-Based Mammogram Classification Models
Sep 26, 2022Abstract:State-of-the-art (SOTA) deep learning mammogram classifiers, trained with weakly-labelled images, often rely on global models that produce predictions with limited interpretability, which is a key barrier to their successful translation into clinical practice. On the other hand, prototype-based models improve interpretability by associating predictions with training image prototypes, but they are less accurate than global models and their prototypes tend to have poor diversity. We address these two issues with the proposal of BRAIxProtoPNet++, which adds interpretability to a global model by ensembling it with a prototype-based model. BRAIxProtoPNet++ distills the knowledge of the global model when training the prototype-based model with the goal of increasing the classification accuracy of the ensemble. Moreover, we propose an approach to increase prototype diversity by guaranteeing that all prototypes are associated with different training images. Experiments on weakly-labelled private and public datasets show that BRAIxProtoPNet++ has higher classification accuracy than SOTA global and prototype-based models. Using lesion localisation to assess model interpretability, we show BRAIxProtoPNet++ is more effective than other prototype-based models and post-hoc explanation of global models. Finally, we show that the diversity of the prototypes learned by BRAIxProtoPNet++ is superior to SOTA prototype-based approaches.
Multi-view Local Co-occurrence and Global Consistency Learning Improve Mammogram Classification Generalisation
Sep 21, 2022Abstract:When analysing screening mammograms, radiologists can naturally process information across two ipsilateral views of each breast, namely the cranio-caudal (CC) and mediolateral-oblique (MLO) views. These multiple related images provide complementary diagnostic information and can improve the radiologist's classification accuracy. Unfortunately, most existing deep learning systems, trained with globally-labelled images, lack the ability to jointly analyse and integrate global and local information from these multiple views. By ignoring the potentially valuable information present in multiple images of a screening episode, one limits the potential accuracy of these systems. Here, we propose a new multi-view global-local analysis method that mimics the radiologist's reading procedure, based on a global consistency learning and local co-occurrence learning of ipsilateral views in mammograms. Extensive experiments show that our model outperforms competing methods, in terms of classification accuracy and generalisation, on a large-scale private dataset and two publicly available datasets, where models are exclusively trained and tested with global labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge