David C. Kale
Effective Representations of Clinical Notes
Aug 16, 2018



Abstract:Clinical notes are a rich source of information about patient state. However, using them to predict clinical events with machine learning models is challenging. They are very high dimensional, sparse and have complex structure. Furthermore, training data is often scarce because it is expensive to obtain reliable labels for many clinical events. These difficulties have traditionally been addressed by manual feature engineering encoding task specific domain knowledge. We explored the use of neural networks and transfer learning to learn representations of clinical notes that are useful for predicting future clinical events of interest, such as all causes mortality, inpatient admissions, and emergency room visits. Our data comprised 2.7 million notes and 115 thousand patients at Stanford Hospital. We used the learned representations, along with commonly used bag of words and topic model representations, as features for predictive models of clinical events. We evaluated the effectiveness of these representations with respect to the performance of the models trained on small datasets. Models using the neural network derived representations performed significantly better than models using the baseline representations with small ($N < 1000$) training datasets. The learned representations offer significant performance gains over commonly used baseline representations for a range of predictive modeling tasks and cohort sizes, offering an effective alternative to task specific feature engineering when plentiful labeled training data is not available.
Multitask Learning and Benchmarking with Clinical Time Series Data
Mar 22, 2017
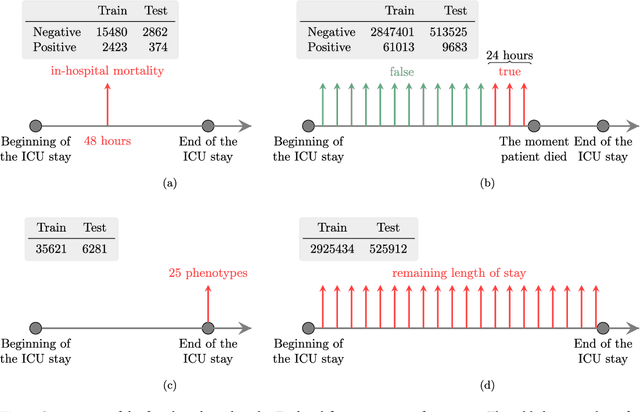
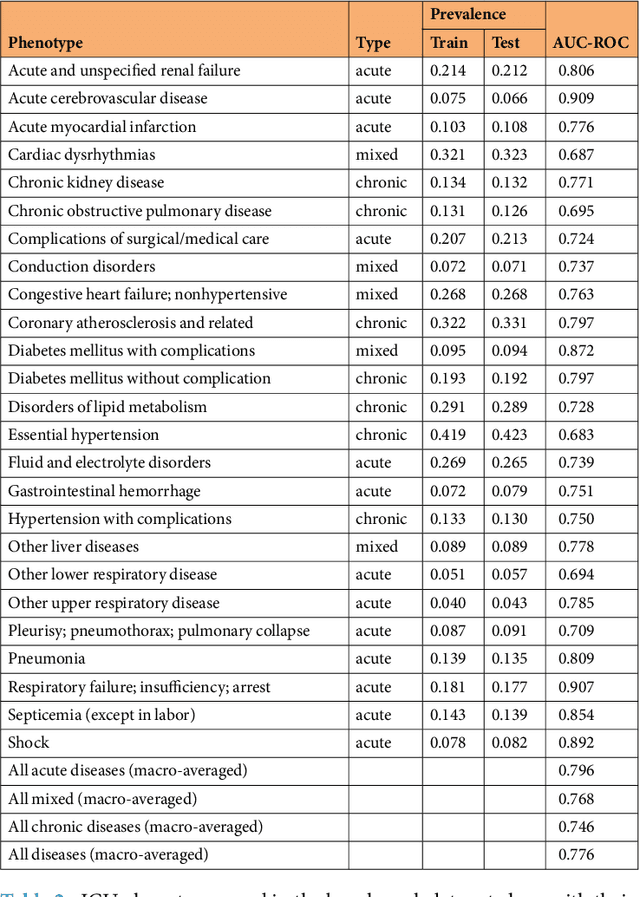

Abstract:Health care is one of the most exciting frontiers in data mining and machine learning. Successful adoption of electronic health records (EHRs) created an explosion in digital clinical data available for analysis, but progress in machine learning for healthcare research has been difficult to measure because of the absence of publicly available benchmark data sets. To address this problem, we propose four clinical prediction benchmarks using data derived from the publicly available Medical Information Mart for Intensive Care (MIMIC-III) database. These tasks cover a range of clinical problems including modeling risk of mortality, forecasting length of stay, detecting physiologic decline, and phenotype classification. We formulate a heterogeneous multitask problem where the goal is to jointly learn multiple clinically relevant prediction tasks based on the same time series data. To address this problem, we propose a novel recurrent neural network (RNN) architecture that leverages the correlations between the various tasks to learn a better predictive model. We validate the proposed neural architecture on this benchmark, and demonstrate that it outperforms strong baselines, including single task RNNs.
Learning to Diagnose with LSTM Recurrent Neural Networks
Mar 21, 2017



Abstract:Clinical medical data, especially in the intensive care unit (ICU), consist of multivariate time series of observations. For each patient visit (or episode), sensor data and lab test results are recorded in the patient's Electronic Health Record (EHR). While potentially containing a wealth of insights, the data is difficult to mine effectively, owing to varying length, irregular sampling and missing data. Recurrent Neural Networks (RNNs), particularly those using Long Short-Term Memory (LSTM) hidden units, are powerful and increasingly popular models for learning from sequence data. They effectively model varying length sequences and capture long range dependencies. We present the first study to empirically evaluate the ability of LSTMs to recognize patterns in multivariate time series of clinical measurements. Specifically, we consider multilabel classification of diagnoses, training a model to classify 128 diagnoses given 13 frequently but irregularly sampled clinical measurements. First, we establish the effectiveness of a simple LSTM network for modeling clinical data. Then we demonstrate a straightforward and effective training strategy in which we replicate targets at each sequence step. Trained only on raw time series, our models outperform several strong baselines, including a multilayer perceptron trained on hand-engineered features.
Phenotyping of Clinical Time Series with LSTM Recurrent Neural Networks
Mar 21, 2017
Abstract:We present a novel application of LSTM recurrent neural networks to multilabel classification of diagnoses given variable-length time series of clinical measurements. Our method outperforms a strong baseline on a variety of metrics.
Intra-day Activity Better Predicts Chronic Conditions
Dec 04, 2016
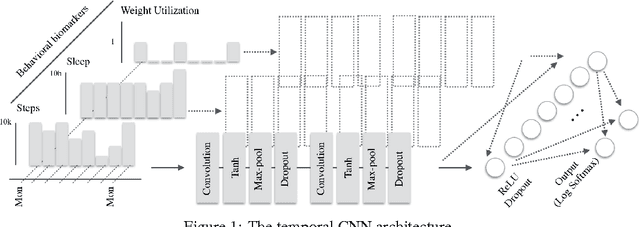
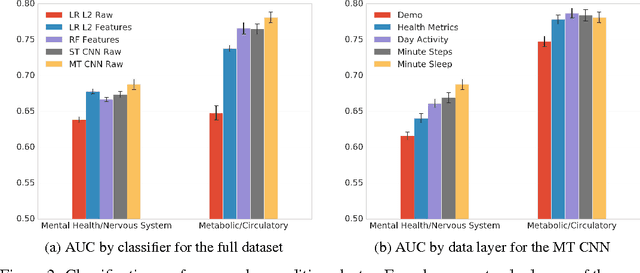
Abstract:In this work we investigate intra-day patterns of activity on a population of 7,261 users of mobile health wearable devices and apps. We show that: (1) using intra-day step and sleep data recorded from passive trackers significantly improves classification performance on self-reported chronic conditions related to mental health and nervous system disorders, (2) Convolutional Neural Networks achieve top classification performance vs. baseline models when trained directly on multivariate time series of activity data, and (3) jointly predicting all condition classes via multi-task learning can be leveraged to extract features that generalize across data sets and achieve the highest classification performance.
Modeling Missing Data in Clinical Time Series with RNNs
Nov 11, 2016



Abstract:We demonstrate a simple strategy to cope with missing data in sequential inputs, addressing the task of multilabel classification of diagnoses given clinical time series. Collected from the pediatric intensive care unit (PICU) at Children's Hospital Los Angeles, our data consists of multivariate time series of observations. The measurements are irregularly spaced, leading to missingness patterns in temporally discretized sequences. While these artifacts are typically handled by imputation, we achieve superior predictive performance by treating the artifacts as features. Unlike linear models, recurrent neural networks can realize this improvement using only simple binary indicators of missingness. For linear models, we show an alternative strategy to capture this signal. Training models on missingness patterns only, we show that for some diseases, what tests are run can be as predictive as the results themselves.
Toward Interpretable Topic Discovery via Anchored Correlation Explanation
Jun 22, 2016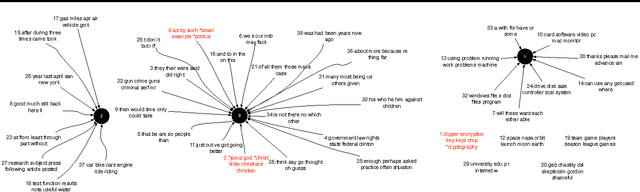
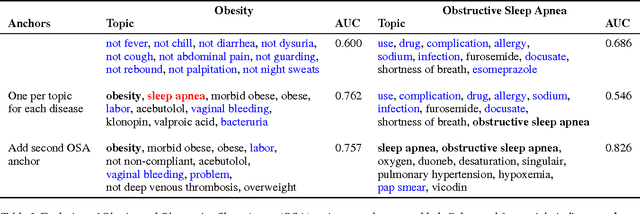


Abstract:Many predictive tasks, such as diagnosing a patient based on their medical chart, are ultimately defined by the decisions of human experts. Unfortunately, encoding experts' knowledge is often time consuming and expensive. We propose a simple way to use fuzzy and informal knowledge from experts to guide discovery of interpretable latent topics in text. The underlying intuition of our approach is that latent factors should be informative about both correlations in the data and a set of relevance variables specified by an expert. Mathematically, this approach is a combination of the information bottleneck and Total Correlation Explanation (CorEx). We give a preliminary evaluation of Anchored CorEx, showing that it produces more coherent and interpretable topics on two distinct corpora.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge