Randall Wetzel
Improving Recurrent Neural Network Responsiveness to Acute Clinical Events
Jul 28, 2020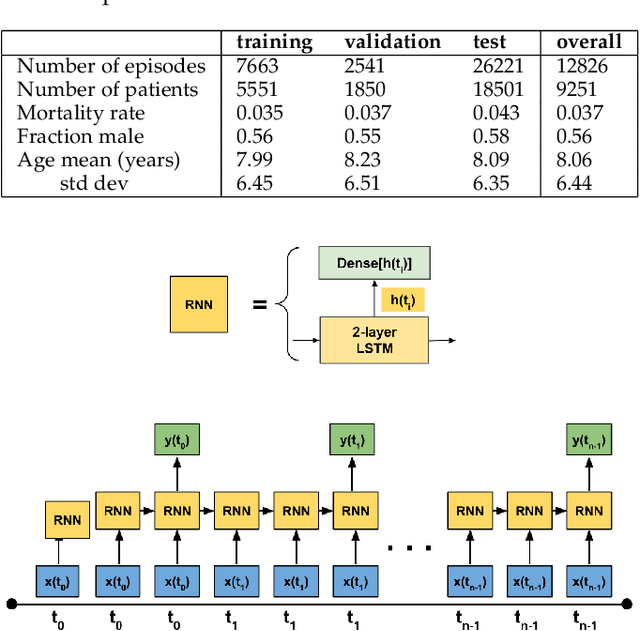
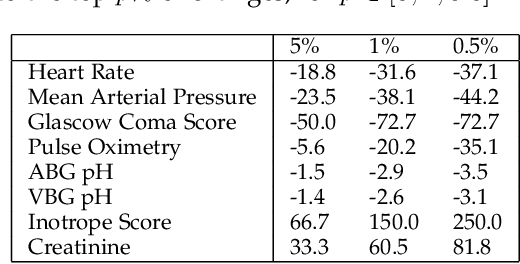
Abstract:Predictive models in acute care settings must be able to immediately recognize precipitous changes in a patient's status when presented with data reflecting such changes. Recurrent neural networks (RNNs) have become common for training and deploying clinical decision support models. They frequently exhibit a delayed response to acute events. New information must propagate through the RNN's cell state memory before the total impact is reflected in the model's predictions. This work presents input data perseveration as a method of training and deploying an RNN model to make its predictions more responsive to newly acquired information: input data is replicated during training and deployment. Each replication of the data input impacts the cell state and output of the RNN, but only the output at the final replication is maintained and broadcast as the prediction for evaluation and deployment purposes. When presented with data reflecting acute events, a model trained and deployed with input perseveration responds with more pronounced immediate changes in predictions and maintains globally robust performance. Such a characteristic is crucial in predictive models for an intensive care unit.
Interpreting a Recurrent Neural Network Model for ICU Mortality Using Learned Binary Masks
Jun 06, 2019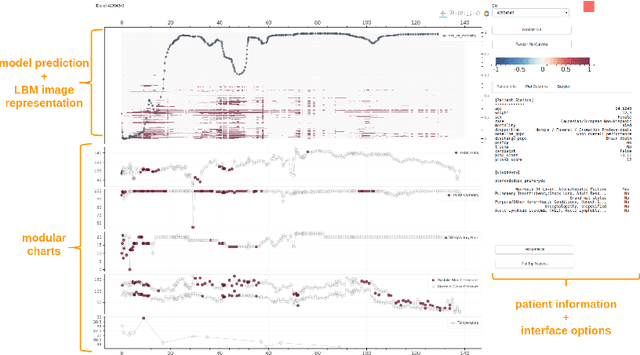
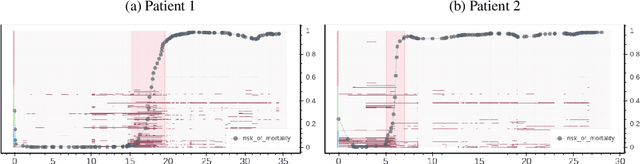
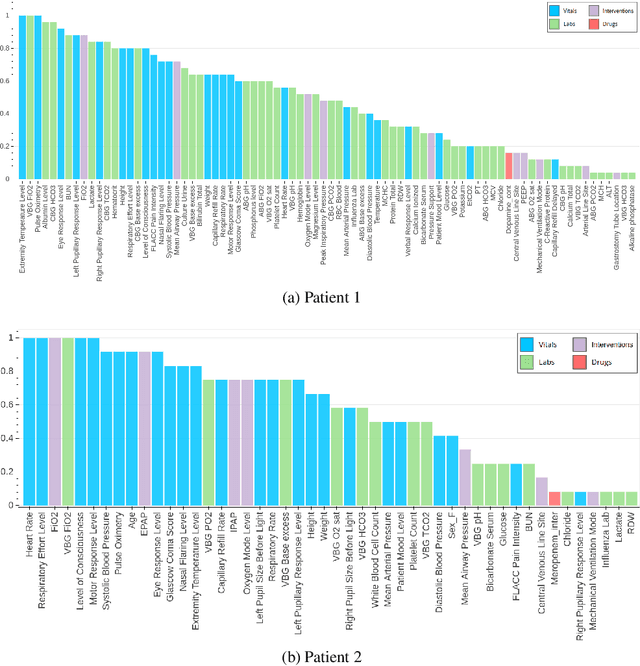
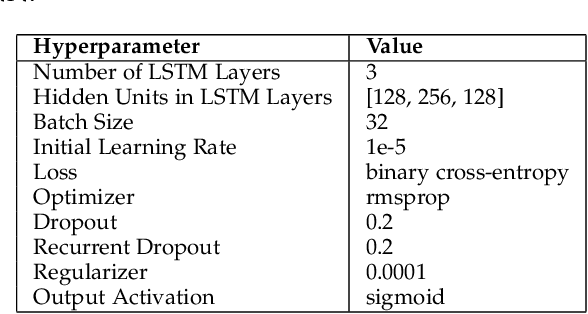
Abstract:An attribution method was developed to interpret a recurrent neural network (RNN) trained to predict a child's risk of ICU mortality using multi-modal, time series data in the Electronic Medical Records. By learning a sparse, binary mask that highlights salient features of the input data, critical features determining an individual patient's severity of illness could be identified. The method, called Learned Binary Masks (LBM), demonstrated that the RNN used different feature sets specific to each patient's illness; and further, the features highlighted aligned with clinical intuition of the patient's disease trajectories. LBM was also used to identify the most salient features across the model, analogous to "feature importance" computed in the Random Forest. This measure of the RNN's feature importance was further used to select the 25% most used features for training a second RNN model. Interestingly, but not surprisingly, the second model maintained similar performance to the model trained on all features. LBM is data-agnostic and can be used to interpret the predictions of any differentiable model.
The Impact of Extraneous Variables on the Performance of Recurrent Neural Network Models in Clinical Tasks
Apr 01, 2019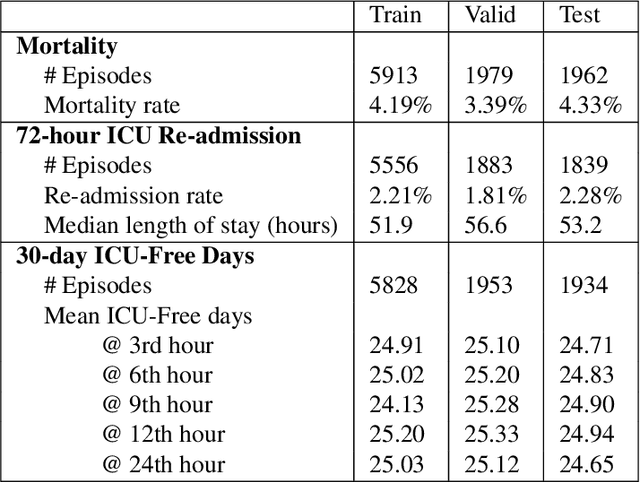
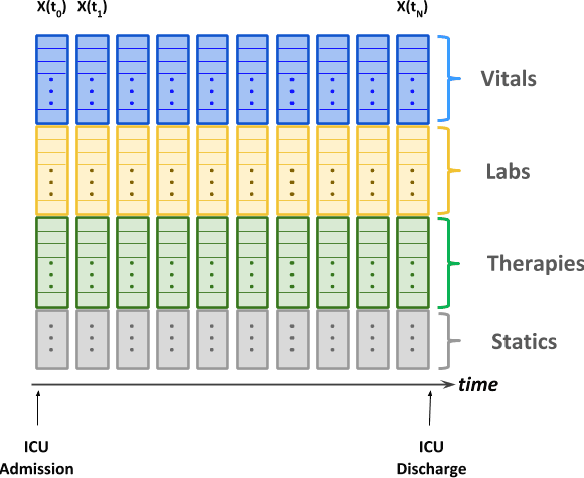
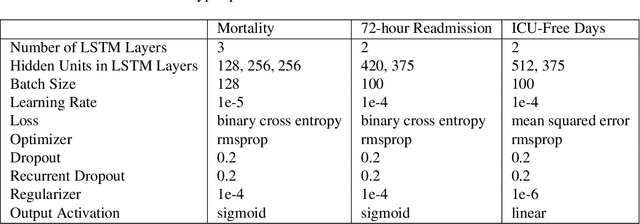
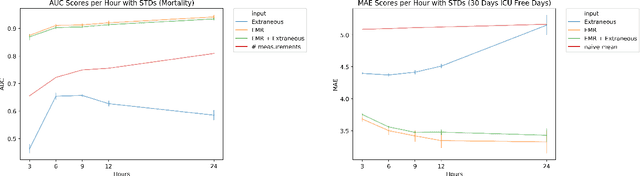
Abstract:Electronic Medical Records (EMR) are a rich source of patient information, including measurements reflecting physiologic signs and administered therapies. Identifying which variables are useful in predicting clinical outcomes can be challenging. Advanced algorithms such as deep neural networks were designed to process high-dimensional inputs containing variables in their measured form, thus bypass separate feature selection or engineering steps. We investigated the effect of extraneous input variables on the predictive performance of Recurrent Neural Networks (RNN) by including in the input vector extraneous variables randomly drawn from theoretical and empirical distributions. RNN models using different input vectors (EMR variables; EMR and extraneous variables; extraneous variables only) were trained to predict three clinical outcomes: in-ICU mortality, 72-hour ICU re-admission, and 30-day ICU-free days. The measured degradations of the RNN's predictive performance with the addition of extraneous variables to EMR variables were negligible.
Predicting Individual Responses to Vasoactive Medications in Children with Septic Shock
Jan 15, 2019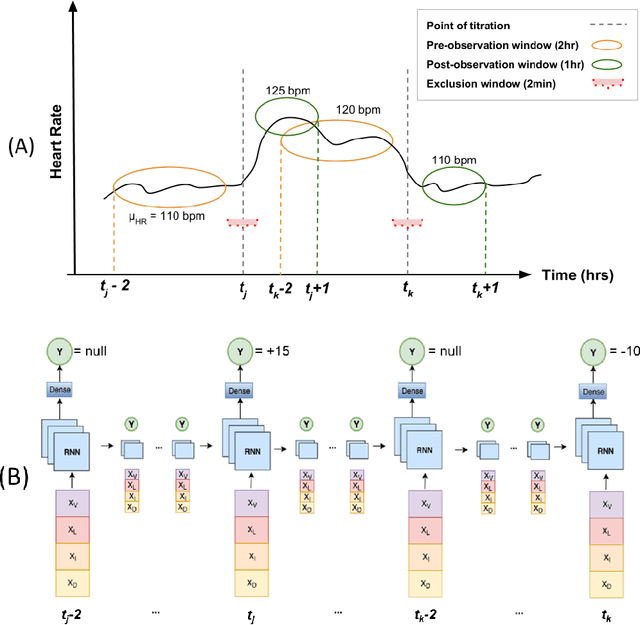
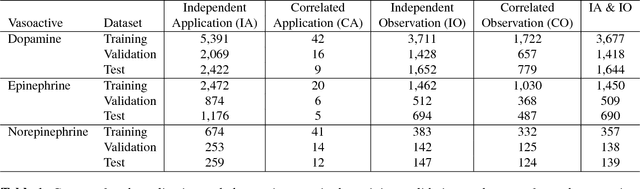
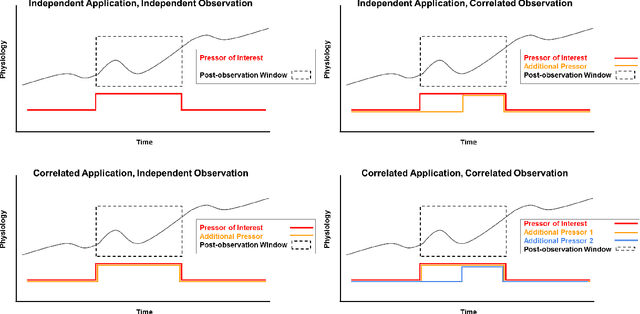
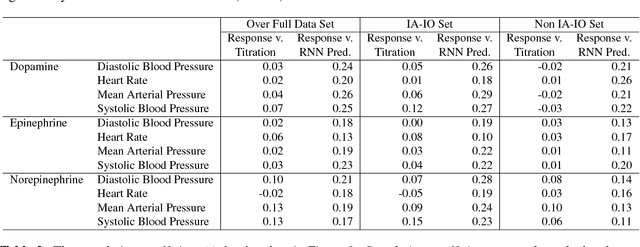
Abstract:Objective: Predict individual septic children's personalized physiologic responses to vasoactive titrations by training a Recurrent Neural Network (RNN) using EMR data. Materials and Methods: This study retrospectively analyzed EMR of patients admitted to a pediatric ICU from 2009 to 2017. Data included charted time series vitals, labs, drugs, and interventions of children with septic shock treated with dopamine, epinephrine, or norepinephrine. A RNN was trained to predict responses in heart rate (HR), systolic blood pressure (SBP), diastolic blood pressure (DBP) and mean arterial pressure (MAP) to 8,640 titrations during 652 septic episodes and evaluated on a holdout set of 3,883 titrations during 254 episodes. A linear regression model using titration data as its sole input was also developed and compared to the RNN model. Evaluation methods included the correlation coefficient between actual physiologic responses and RNN predictions, mean absolute error (MAE), and area under the receiver operating characteristic curve (AUC). Results: The actual physiologic responses displayed significant variability and were more accurately predicted by the RNN model than by titration alone (r=0.20 vs r=0.05, p<0.01). The RNN showed MAE and AUC improvements over the linear model. The RNN's MAEs associated with dopamine and epinephrine were 1-3% lower than the linear regression model MAE for HR, SBP, DBP, and MAP. Across all vitals vasoactives, the RNN achieved 1-19% AUC improvement over the linear model. Conclusion: This initial attempt in pediatric critical care to predict individual physiologic responses to vasoactive dose changes in children with septic shock demonstrated an RNN model showed some improvement over a linear model. While not yet clinically applicable, further development may assist clinical administration of vasoactive medications in children with septic shock.
Predicting Individual Physiologically Acceptable States for Discharge from a Pediatric Intensive Care Unit
Dec 18, 2017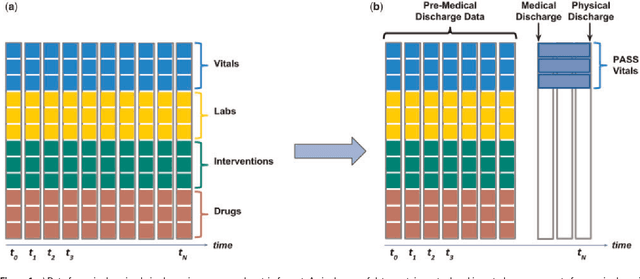
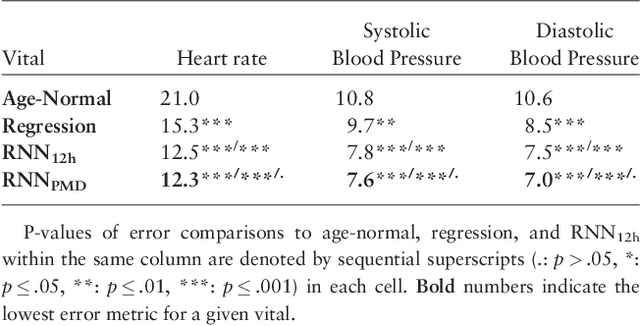
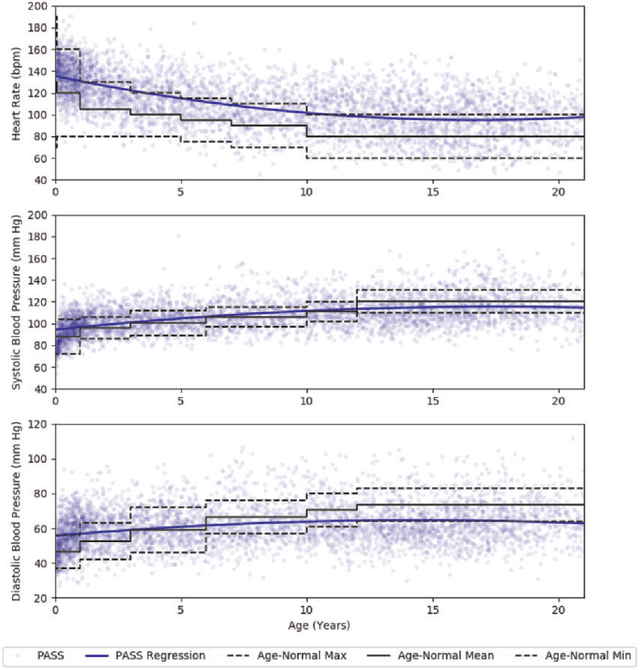
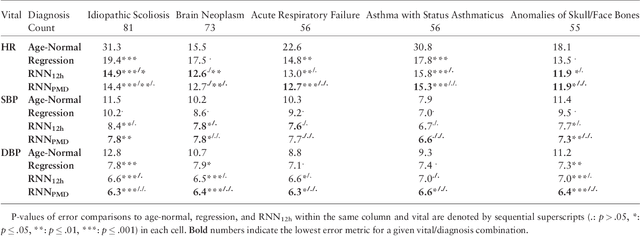
Abstract:Objective: Predict patient-specific vitals deemed medically acceptable for discharge from a pediatric intensive care unit (ICU). Design: The means of each patient's hr, sbp and dbp measurements between their medical and physical discharge from the ICU were computed as a proxy for their physiologically acceptable state space (PASS) for successful ICU discharge. These individual PASS values were compared via root mean squared error (rMSE) to population age-normal vitals, a polynomial regression through the PASS values of a Pediatric ICU (PICU) population and predictions from two recurrent neural network models designed to predict personalized PASS within the first twelve hours following ICU admission. Setting: PICU at Children's Hospital Los Angeles (CHLA). Patients: 6,899 PICU episodes (5,464 patients) collected between 2009 and 2016. Interventions: None. Measurements: Each episode data contained 375 variables representing vitals, labs, interventions, and drugs. They also included a time indicator for PICU medical discharge and physical discharge. Main Results: The rMSEs between individual PASS values and population age-normals (hr: 25.9 bpm, sbp: 13.4 mmHg, dbp: 13.0 mmHg) were larger than the rMSEs corresponding to the polynomial regression (hr: 19.1 bpm, sbp: 12.3 mmHg, dbp: 10.8 mmHg). The rMSEs from the best performing RNN model were the lowest (hr: 16.4 bpm; sbp: 9.9 mmHg, dbp: 9.0 mmHg). Conclusion: PICU patients are a unique subset of the general population, and general age-normal vitals may not be suitable as target values indicating physiologic stability at discharge. Age-normal vitals that were specifically derived from the medical-to-physical discharge window of ICU patients may be more appropriate targets for 'acceptable' physiologic state for critical care patients. Going beyond simple age bins, an RNN model can provide more personalized target values.
The Dependence of Machine Learning on Electronic Medical Record Quality
Mar 23, 2017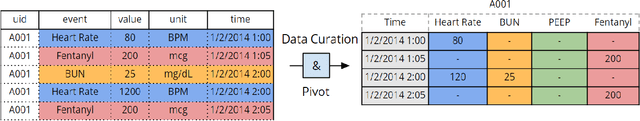
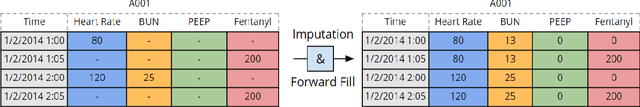
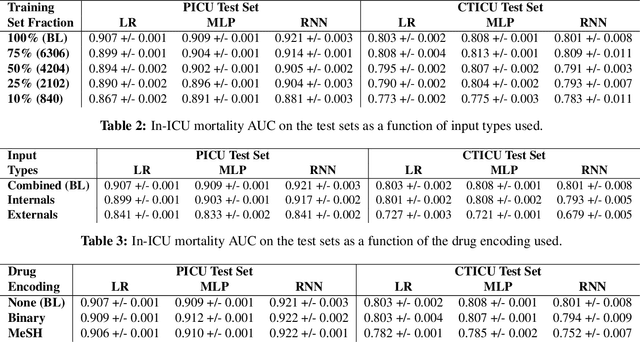
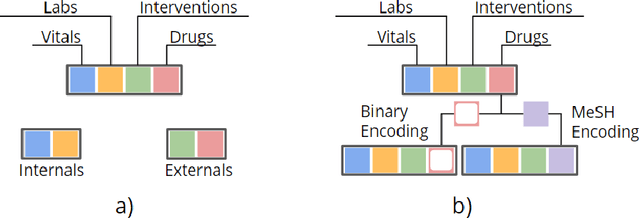
Abstract:There is growing interest in applying machine learning methods to Electronic Medical Records (EMR). Across different institutions, however, EMR quality can vary widely. This work investigated the impact of this disparity on the performance of three advanced machine learning algorithms: logistic regression, multilayer perceptron, and recurrent neural network. The EMR disparity was emulated using different permutations of the EMR collected at Children's Hospital Los Angeles (CHLA) Pediatric Intensive Care Unit (PICU) and Cardiothoracic Intensive Care Unit (CTICU). The algorithms were trained using patients from the PICU to predict in-ICU mortality for patients in a held out set of PICU and CTICU patients. The disparate patient populations between the PICU and CTICU provide an estimate of generalization errors across different ICUs. We quantified and evaluated the generalization of these algorithms on varying EMR size, input types, and fidelity of data.
Learning to Diagnose with LSTM Recurrent Neural Networks
Mar 21, 2017



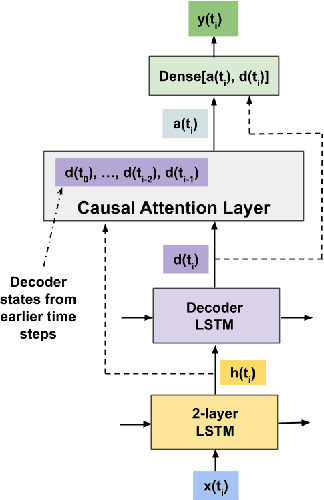
Abstract:Clinical medical data, especially in the intensive care unit (ICU), consist of multivariate time series of observations. For each patient visit (or episode), sensor data and lab test results are recorded in the patient's Electronic Health Record (EHR). While potentially containing a wealth of insights, the data is difficult to mine effectively, owing to varying length, irregular sampling and missing data. Recurrent Neural Networks (RNNs), particularly those using Long Short-Term Memory (LSTM) hidden units, are powerful and increasingly popular models for learning from sequence data. They effectively model varying length sequences and capture long range dependencies. We present the first study to empirically evaluate the ability of LSTMs to recognize patterns in multivariate time series of clinical measurements. Specifically, we consider multilabel classification of diagnoses, training a model to classify 128 diagnoses given 13 frequently but irregularly sampled clinical measurements. First, we establish the effectiveness of a simple LSTM network for modeling clinical data. Then we demonstrate a straightforward and effective training strategy in which we replicate targets at each sequence step. Trained only on raw time series, our models outperform several strong baselines, including a multilayer perceptron trained on hand-engineered features.
Modeling Missing Data in Clinical Time Series with RNNs
Nov 11, 2016



Abstract:We demonstrate a simple strategy to cope with missing data in sequential inputs, addressing the task of multilabel classification of diagnoses given clinical time series. Collected from the pediatric intensive care unit (PICU) at Children's Hospital Los Angeles, our data consists of multivariate time series of observations. The measurements are irregularly spaced, leading to missingness patterns in temporally discretized sequences. While these artifacts are typically handled by imputation, we achieve superior predictive performance by treating the artifacts as features. Unlike linear models, recurrent neural networks can realize this improvement using only simple binary indicators of missingness. For linear models, we show an alternative strategy to capture this signal. Training models on missingness patterns only, we show that for some diseases, what tests are run can be as predictive as the results themselves.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge