David A. Coomes
Unsupervised detection of ash dieback disease (Hymenoscyphus fraxineus) using diffusion-based hyperspectral image clustering
Apr 19, 2022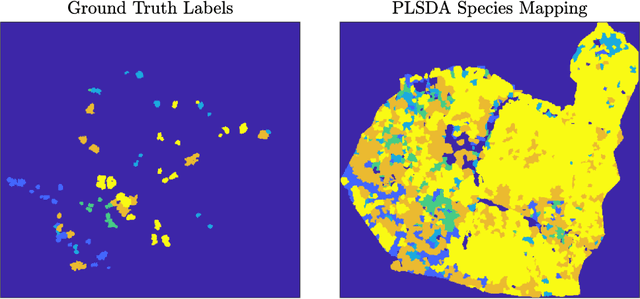
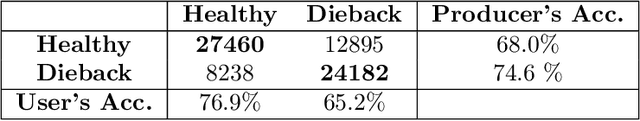
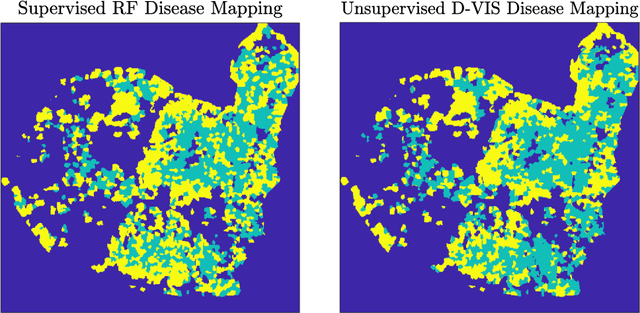
Abstract:Ash dieback (Hymenoscyphus fraxineus) is an introduced fungal disease that is causing the widespread death of ash trees across Europe. Remote sensing hyperspectral images encode rich structure that has been exploited for the detection of dieback disease in ash trees using supervised machine learning techniques. However, to understand the state of forest health at landscape-scale, accurate unsupervised approaches are needed. This article investigates the use of the unsupervised Diffusion and VCA-Assisted Image Segmentation (D-VIS) clustering algorithm for the detection of ash dieback disease in a forest site near Cambridge, United Kingdom. The unsupervised clustering presented in this work has high overlap with the supervised classification of previous work on this scene (overall accuracy = 71%). Thus, unsupervised learning may be used for the remote detection of ash dieback disease without the need for expert labeling.
SLIC-UAV: A Method for monitoring recovery in tropical restoration projects through identification of signature species using UAVs
Jun 11, 2020

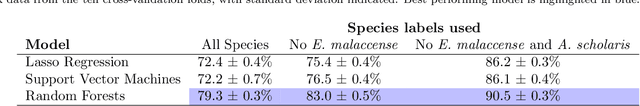
Abstract:Logged forests cover four million square kilometres of the tropics and restoring these forests is essential if we are to avoid the worst impacts of climate change, yet monitoring recovery is challenging. Tracking the abundance of visually identifiable, early-successional species enables successional status and thereby restoration progress to be evaluated. Here we present a new pipeline, SLIC-UAV, for processing Unmanned Aerial Vehicle (UAV) imagery to map early-successional species in tropical forests. The pipeline is novel because it comprises: (a) a time-efficient approach for labelling crowns from UAV imagery; (b) machine learning of species based on spectral and textural features within individual tree crowns, and (c) automatic segmentation of orthomosaiced UAV imagery into 'superpixels', using Simple Linear Iterative Clustering (SLIC). Creating superpixels reduces the dataset's dimensionality and focuses prediction onto clusters of pixels, greatly improving accuracy. To demonstrate SLIC-UAV, support vector machines and random forests were used to predict the species of hand-labelled crowns in a restoration concession in Indonesia. Random forests were most accurate at discriminating species for whole crowns, with accuracy ranging from 79.3% when mapping five common species, to 90.5% when mapping the three most visually-distinctive species. In contrast, support vector machines proved better for labelling automatically segmented superpixels, with accuracy ranging from 74.3% to 91.7% for the same species. Models were extended to map species across 100 hectares of forest. The study demonstrates the power of SLIC-UAV for mapping characteristic early-successional tree species as an indicator of successional stage within tropical forest restoration areas. Continued effort is needed to develop easy-to-implement and low-cost technology to improve the affordability of project management.
Three-dimensional Segmentation of Trees Through a Flexible Multi-Class Graph Cut Algorithm (MCGC)
Mar 20, 2019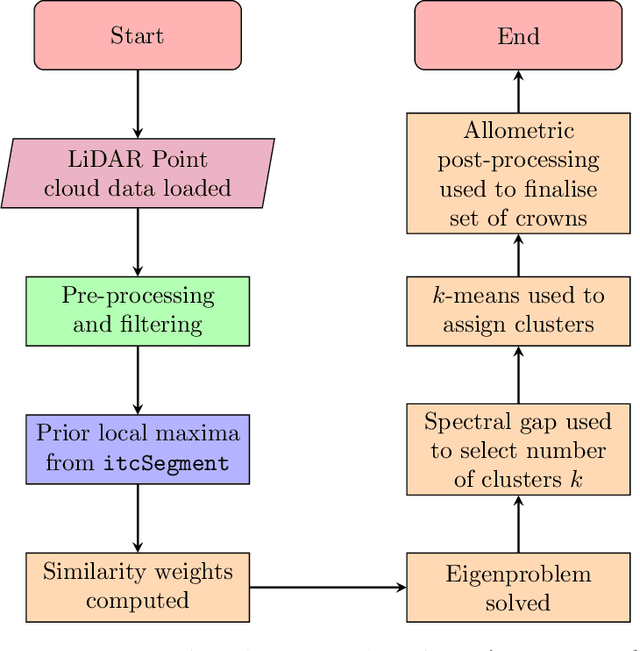
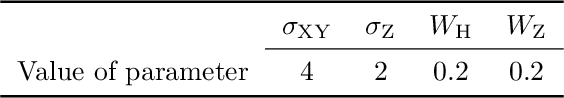
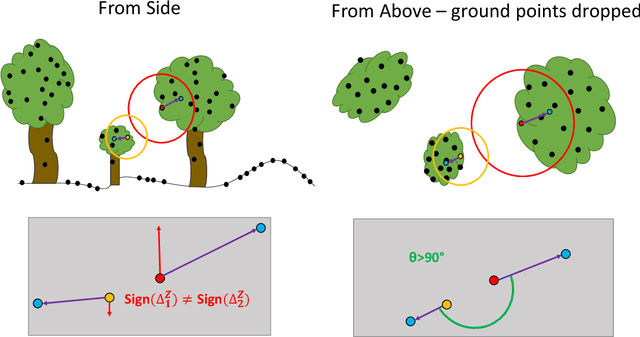
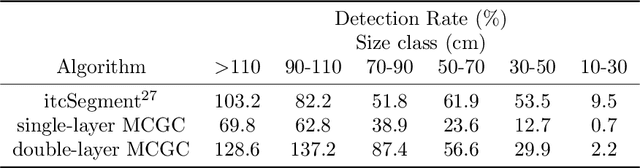
Abstract:Developing a robust algorithm for automatic individual tree crown (ITC) detection from laser scanning datasets is important for tracking the responses of trees to anthropogenic change. Such approaches allow the size, growth and mortality of individual trees to be measured, enabling forest carbon stocks and dynamics to be tracked and understood. Many algorithms exist for structurally simple forests including coniferous forests and plantations. Finding a robust solution for structurally complex, species-rich tropical forests remains a challenge; existing segmentation algorithms often perform less well than simple area-based approaches when estimating plot-level biomass. Here we describe a Multi-Class Graph Cut (MCGC) approach to tree crown delineation. This uses local three-dimensional geometry and density information, alongside knowledge of crown allometries, to segment individual tree crowns from LiDAR point clouds. Our approach robustly identifies trees in the top and intermediate layers of the canopy, but cannot recognise small trees. From these three-dimensional crowns, we are able to measure individual tree biomass. Comparing these estimates to those from permanent inventory plots, our algorithm is able to produce robust estimates of hectare-scale carbon density, demonstrating the power of ITC approaches in monitoring forests. The flexibility of our method to add additional dimensions of information, such as spectral reflectance, make this approach an obvious avenue for future development and extension to other sources of three-dimensional data, such as structure from motion datasets.
Blind Image Fusion for Hyperspectral Imaging with the Directional Total Variation
Apr 09, 2018



Abstract:Hyperspectral imaging is a cutting-edge type of remote sensing used for mapping vegetation properties, rock minerals and other materials. A major drawback of hyperspectral imaging devices is their intrinsic low spatial resolution. In this paper, we propose a method for increasing the spatial resolution of a hyperspectral image by fusing it with an image of higher spatial resolution that was obtained with a different imaging modality. This is accomplished by solving a variational problem in which the regularization functional is the directional total variation. To accommodate for possible mis-registrations between the two images, we consider a non-convex blind super-resolution problem where both a fused image and the corresponding convolution kernel are estimated. Using this approach, our model can realign the given images if needed. Our experimental results indicate that the non-convexity is negligible in practice and that reliable solutions can be computed using a variety of different optimization algorithms. Numerical results on real remote sensing data from plant sciences and urban monitoring show the potential of the proposed method and suggests that it is robust with respect to the regularization parameters, mis-registration and the shape of the kernel.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge