Dane Bell
Sieve-based Coreference Resolution in the Biomedical Domain
Sep 02, 2016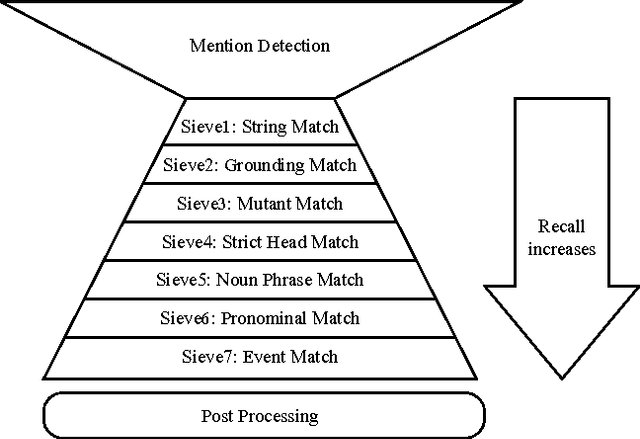


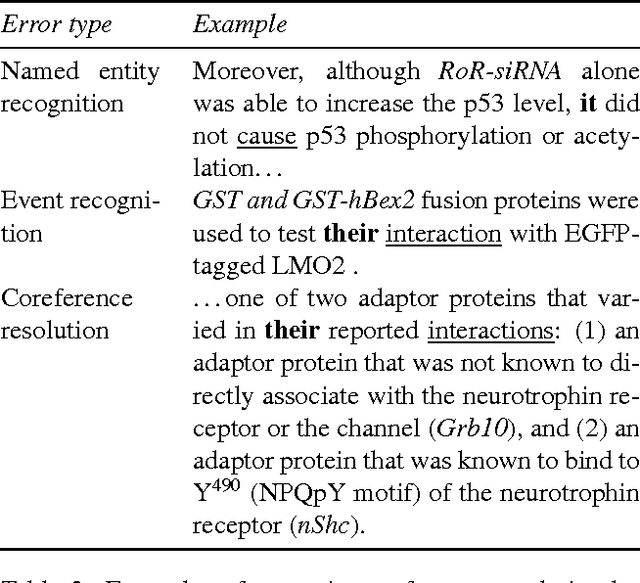
Abstract:We describe challenges and advantages unique to coreference resolution in the biomedical domain, and a sieve-based architecture that leverages domain knowledge for both entity and event coreference resolution. Domain-general coreference resolution algorithms perform poorly on biomedical documents, because the cues they rely on such as gender are largely absent in this domain, and because they do not encode domain-specific knowledge such as the number and type of participants required in chemical reactions. Moreover, it is difficult to directly encode this knowledge into most coreference resolution algorithms because they are not rule-based. Our rule-based architecture uses sequentially applied hand-designed "sieves", with the output of each sieve informing and constraining subsequent sieves. This architecture provides a 3.2% increase in throughput to our Reach event extraction system with precision parallel to that of the stricter system that relies solely on syntactic patterns for extraction.
SnapToGrid: From Statistical to Interpretable Models for Biomedical Information Extraction
Jun 30, 2016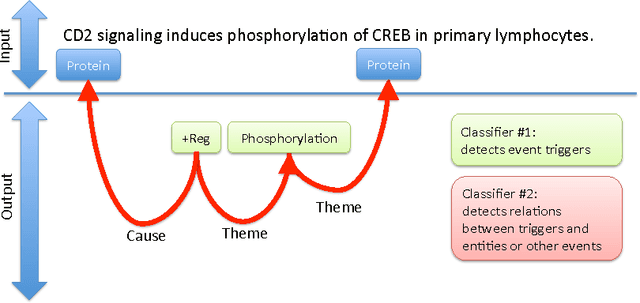
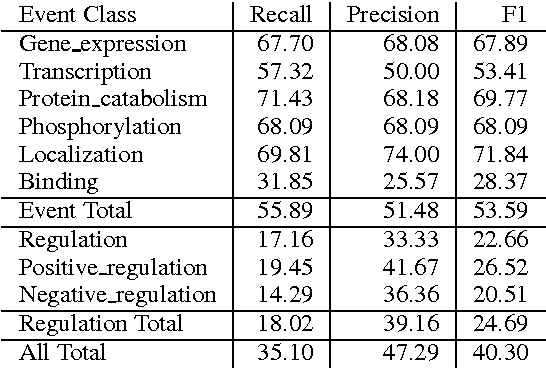
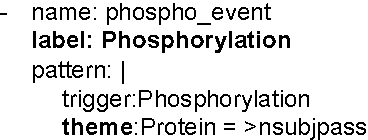
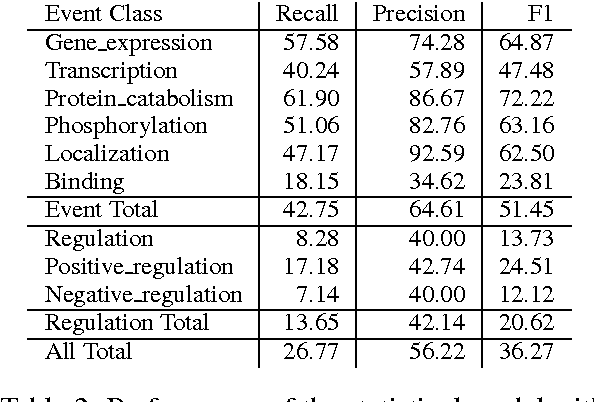
Abstract:We propose an approach for biomedical information extraction that marries the advantages of machine learning models, e.g., learning directly from data, with the benefits of rule-based approaches, e.g., interpretability. Our approach starts by training a feature-based statistical model, then converts this model to a rule-based variant by converting its features to rules, and "snapping to grid" the feature weights to discrete votes. In doing so, our proposal takes advantage of the large body of work in machine learning, but it produces an interpretable model, which can be directly edited by experts. We evaluate our approach on the BioNLP 2009 event extraction task. Our results show that there is a small performance penalty when converting the statistical model to rules, but the gain in interpretability compensates for that: with minimal effort, human experts improve this model to have similar performance to the statistical model that served as starting point.
This before That: Causal Precedence in the Biomedical Domain
Jun 26, 2016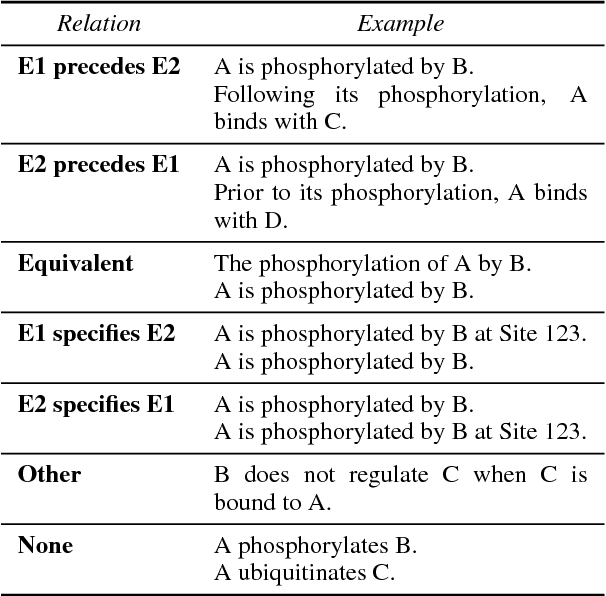
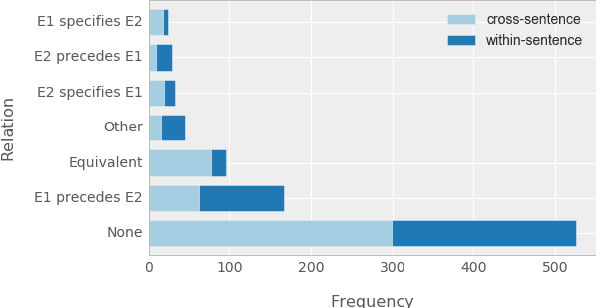
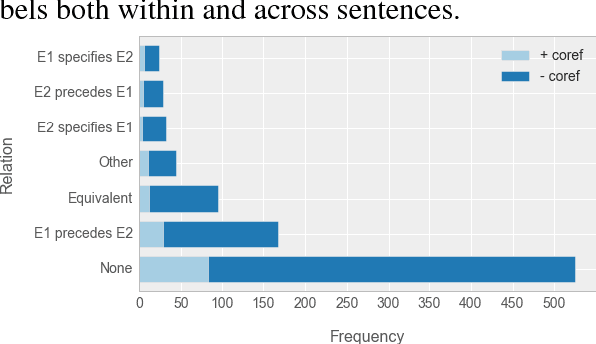
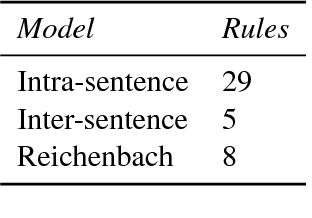
Abstract:Causal precedence between biochemical interactions is crucial in the biomedical domain, because it transforms collections of individual interactions, e.g., bindings and phosphorylations, into the causal mechanisms needed to inform meaningful search and inference. Here, we analyze causal precedence in the biomedical domain as distinct from open-domain, temporal precedence. First, we describe a novel, hand-annotated text corpus of causal precedence in the biomedical domain. Second, we use this corpus to investigate a battery of models of precedence, covering rule-based, feature-based, and latent representation models. The highest-performing individual model achieved a micro F1 of 43 points, approaching the best performers on the simpler temporal-only precedence tasks. Feature-based and latent representation models each outperform the rule-based models, but their performance is complementary to one another. We apply a sieve-based architecture to capitalize on this lack of overlap, achieving a micro F1 score of 46 points.
Towards using social media to identify individuals at risk for preventable chronic illness
Mar 11, 2016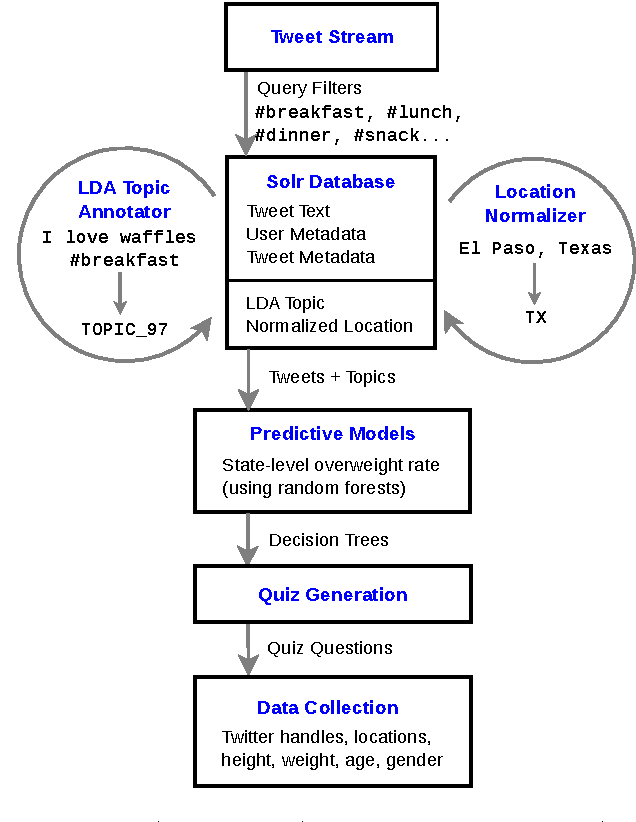

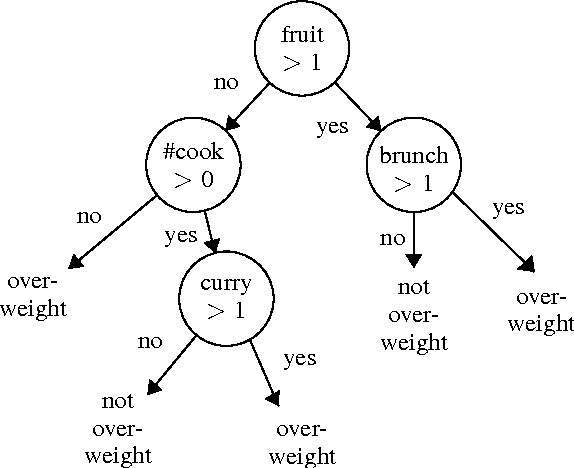
Abstract:We describe a strategy for the acquisition of training data necessary to build a social-media-driven early detection system for individuals at risk for (preventable) type 2 diabetes mellitus (T2DM). The strategy uses a game-like quiz with data and questions acquired semi-automatically from Twitter. The questions are designed to inspire participant engagement and collect relevant data to train a public-health model applied to individuals. Prior systems designed to use social media such as Twitter to predict obesity (a risk factor for T2DM) operate on entire communities such as states, counties, or cities, based on statistics gathered by government agencies. Because there is considerable variation among individuals within these groups, training data on the individual level would be more effective, but this data is difficult to acquire. The approach proposed here aims to address this issue. Our strategy has two steps. First, we trained a random forest classifier on data gathered from (public) Twitter statuses and state-level statistics with state-of-the-art accuracy. We then converted this classifier into a 20-questions-style quiz and made it available online. In doing so, we achieved high engagement with individuals that took the quiz, while also building a training set of voluntarily supplied individual-level data for future classification.
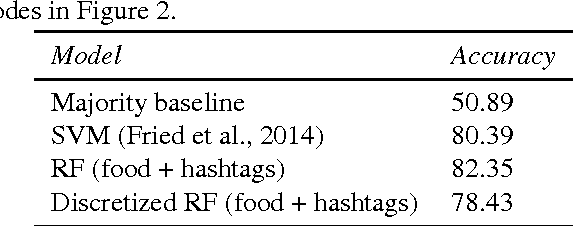
Analyzing the Language of Food on Social Media
Sep 11, 2014
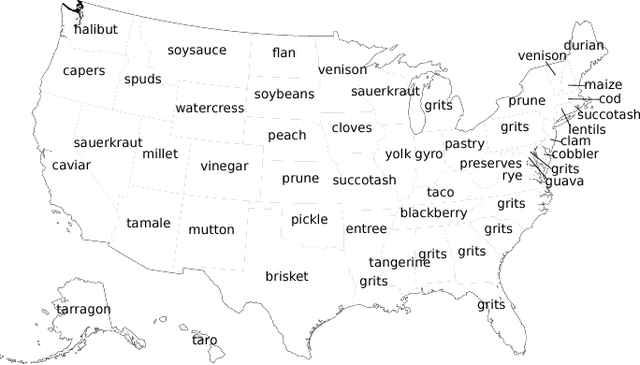

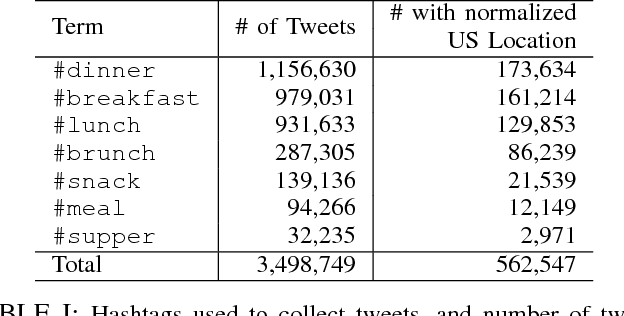
Abstract:We investigate the predictive power behind the language of food on social media. We collect a corpus of over three million food-related posts from Twitter and demonstrate that many latent population characteristics can be directly predicted from this data: overweight rate, diabetes rate, political leaning, and home geographical location of authors. For all tasks, our language-based models significantly outperform the majority-class baselines. Performance is further improved with more complex natural language processing, such as topic modeling. We analyze which textual features have most predictive power for these datasets, providing insight into the connections between the language of food, geographic locale, and community characteristics. Lastly, we design and implement an online system for real-time query and visualization of the dataset. Visualization tools, such as geo-referenced heatmaps, semantics-preserving wordclouds and temporal histograms, allow us to discover more complex, global patterns mirrored in the language of food.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge