Gus Hahn-Powell
Syntax-driven Data Augmentation for Named Entity Recognition
Aug 15, 2022


Abstract:In low resource settings, data augmentation strategies are commonly leveraged to improve performance. Numerous approaches have attempted document-level augmentation (e.g., text classification), but few studies have explored token-level augmentation. Performed naively, data augmentation can produce semantically incongruent and ungrammatical examples. In this work, we compare simple masked language model replacement and an augmentation method using constituency tree mutations to improve the performance of named entity recognition in low-resource settings with the aim of preserving linguistic cohesion of the augmented sentences.
Computer-assisted construct classification of organizational performance concerning different stakeholder groups
Jul 11, 2021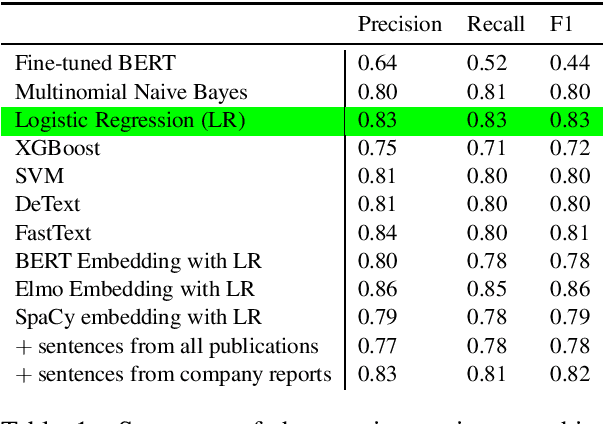
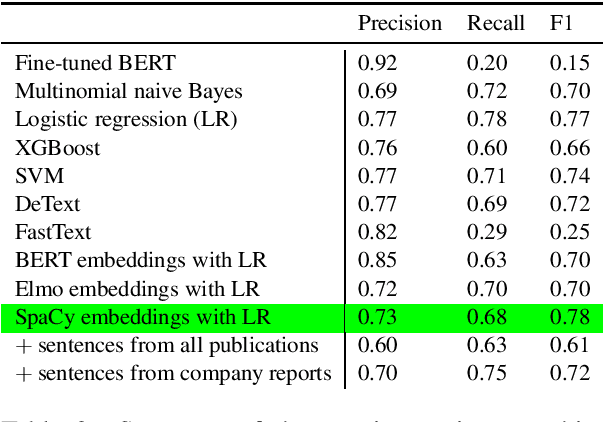
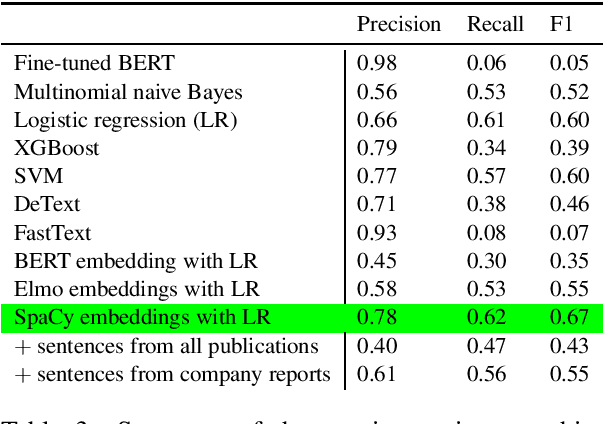
Abstract:The number of research articles in business and management has dramatically increased along with terminology, constructs, and measures. Proper classification of organizational performance constructs from research articles plays an important role in categorizing the literature and understanding to whom its research implications may be relevant. In this work, we classify constructs (i.e., concepts and terminology used to capture different aspects of organizational performance) in research articles into a three-level categorization: (a) performance and non-performance categories (Level 0); (b) for performance constructs, stakeholder group-level of performance concerning investors, customers, employees, and the society (community and natural environment) (Level 1); and (c) for each stakeholder group-level, subcategories of different ways of measurement (Level 2). We observed that increasing contextual information with features extracted from surrounding sentences and external references improves classification of disaggregate-level labels, given limited training data. Our research has implications for computer-assisted construct identification and classification - an essential step for research synthesis.
Text Annotation Graphs: Annotating Complex Natural Language Phenomena
Mar 01, 2018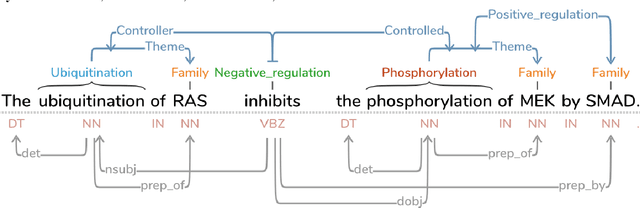
Abstract:This paper introduces a new web-based software tool for annotating text, Text Annotation Graphs, or TAG. It provides functionality for representing complex relationships between words and word phrases that are not available in other software tools, including the ability to define and visualize relationships between the relationships themselves (semantic hypergraphs). Additionally, we include an approach to representing text annotations in which annotation subgraphs, or semantic summaries, are used to show relationships outside of the sequential context of the text itself. Users can use these subgraphs to quickly find similar structures within the current document or external annotated documents. Initially, TAG was developed to support information extraction tasks on a large database of biomedical articles. However, our software is flexible enough to support a wide range of annotation tasks for any domain. Examples are provided that showcase TAG's capabilities on morphological parsing and event extraction tasks. The TAG software is available at: https://github.com/ CreativeCodingLab/TextAnnotationGraphs.
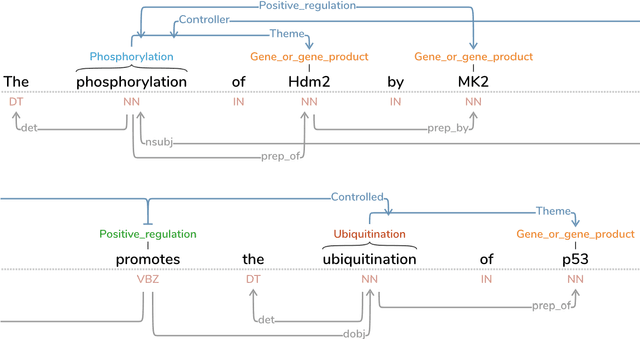
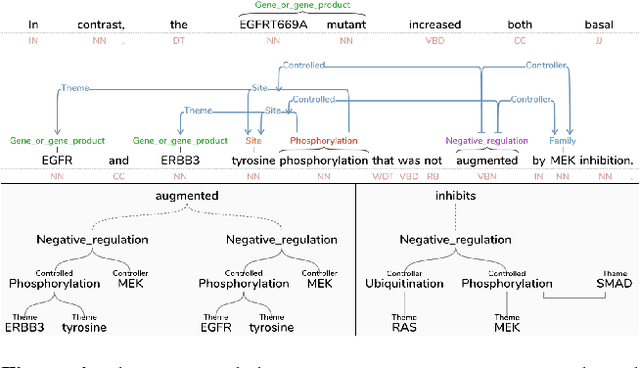
Sieve-based Coreference Resolution in the Biomedical Domain
Sep 02, 2016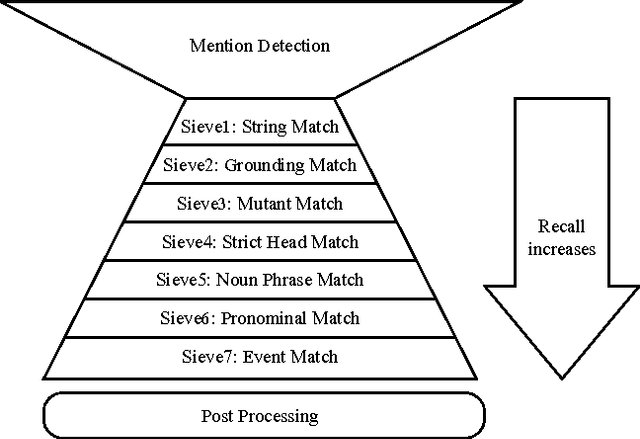


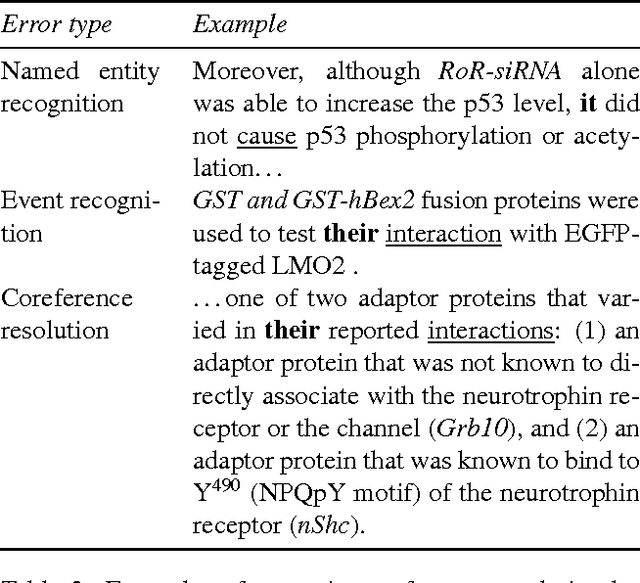
Abstract:We describe challenges and advantages unique to coreference resolution in the biomedical domain, and a sieve-based architecture that leverages domain knowledge for both entity and event coreference resolution. Domain-general coreference resolution algorithms perform poorly on biomedical documents, because the cues they rely on such as gender are largely absent in this domain, and because they do not encode domain-specific knowledge such as the number and type of participants required in chemical reactions. Moreover, it is difficult to directly encode this knowledge into most coreference resolution algorithms because they are not rule-based. Our rule-based architecture uses sequentially applied hand-designed "sieves", with the output of each sieve informing and constraining subsequent sieves. This architecture provides a 3.2% increase in throughput to our Reach event extraction system with precision parallel to that of the stricter system that relies solely on syntactic patterns for extraction.
SnapToGrid: From Statistical to Interpretable Models for Biomedical Information Extraction
Jun 30, 2016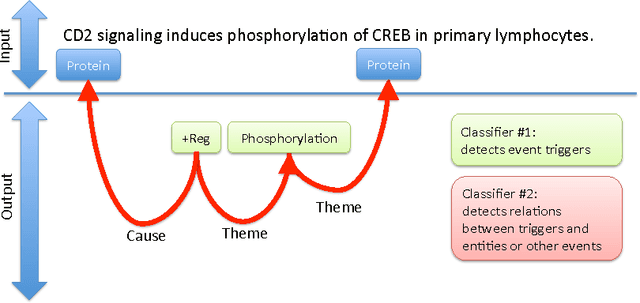
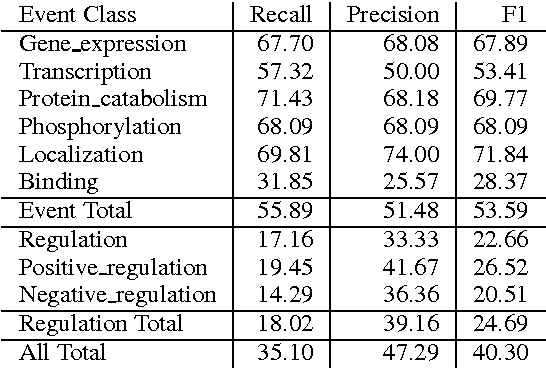
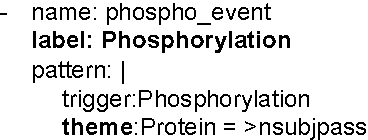
Abstract:We propose an approach for biomedical information extraction that marries the advantages of machine learning models, e.g., learning directly from data, with the benefits of rule-based approaches, e.g., interpretability. Our approach starts by training a feature-based statistical model, then converts this model to a rule-based variant by converting its features to rules, and "snapping to grid" the feature weights to discrete votes. In doing so, our proposal takes advantage of the large body of work in machine learning, but it produces an interpretable model, which can be directly edited by experts. We evaluate our approach on the BioNLP 2009 event extraction task. Our results show that there is a small performance penalty when converting the statistical model to rules, but the gain in interpretability compensates for that: with minimal effort, human experts improve this model to have similar performance to the statistical model that served as starting point.
This before That: Causal Precedence in the Biomedical Domain
Jun 26, 2016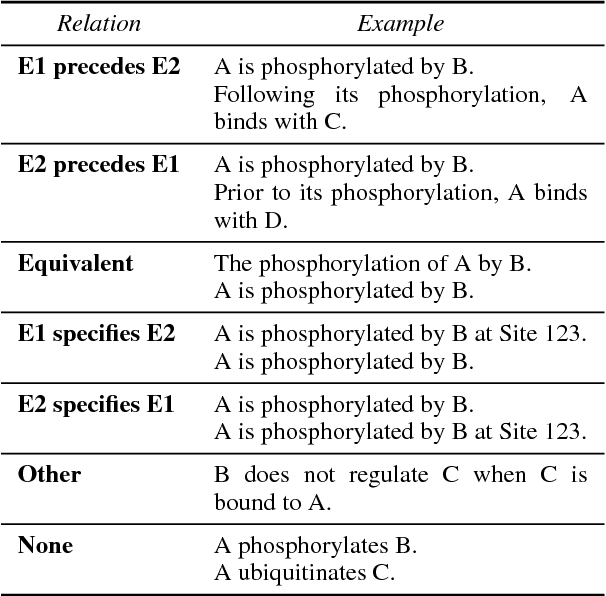
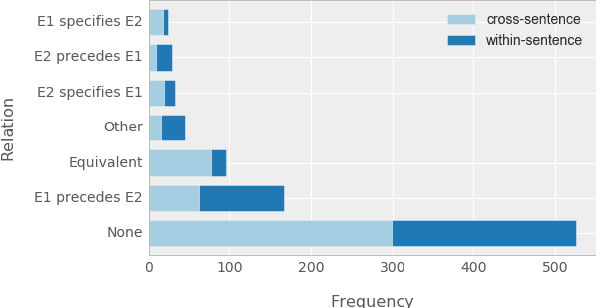
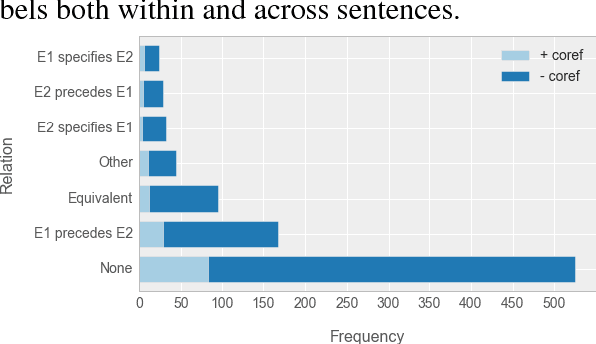
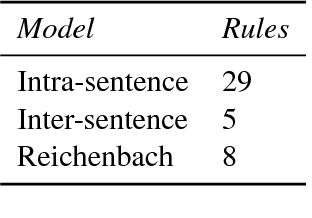
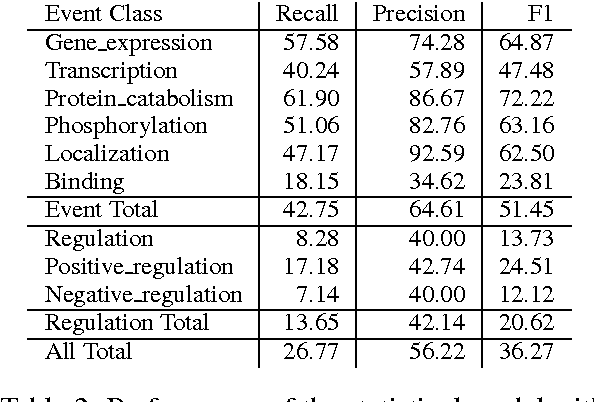
Abstract:Causal precedence between biochemical interactions is crucial in the biomedical domain, because it transforms collections of individual interactions, e.g., bindings and phosphorylations, into the causal mechanisms needed to inform meaningful search and inference. Here, we analyze causal precedence in the biomedical domain as distinct from open-domain, temporal precedence. First, we describe a novel, hand-annotated text corpus of causal precedence in the biomedical domain. Second, we use this corpus to investigate a battery of models of precedence, covering rule-based, feature-based, and latent representation models. The highest-performing individual model achieved a micro F1 of 43 points, approaching the best performers on the simpler temporal-only precedence tasks. Feature-based and latent representation models each outperform the rule-based models, but their performance is complementary to one another. We apply a sieve-based architecture to capitalize on this lack of overlap, achieving a micro F1 score of 46 points.
Description of the Odin Event Extraction Framework and Rule Language
Sep 24, 2015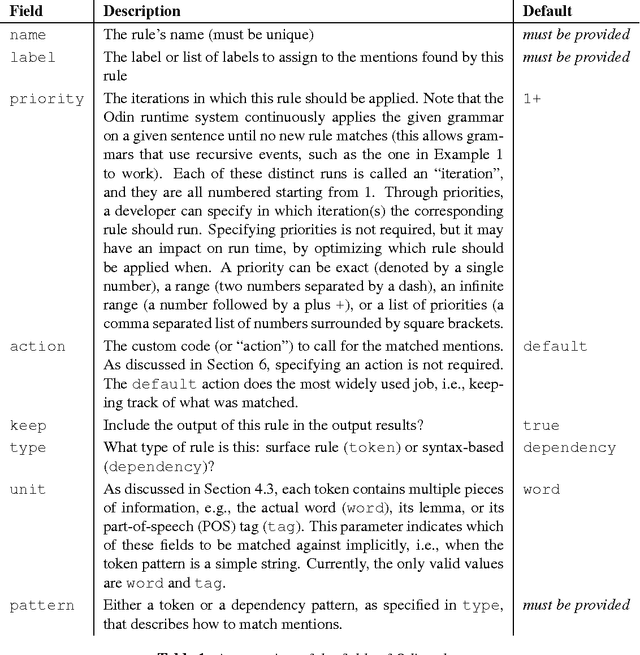
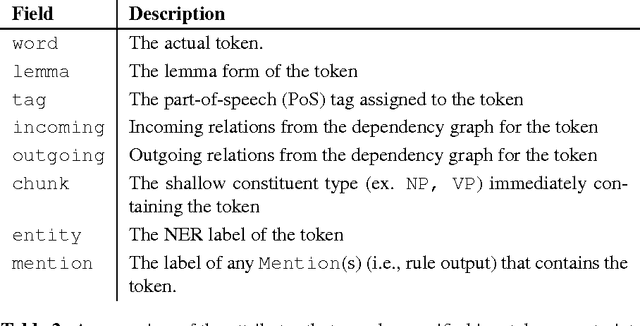
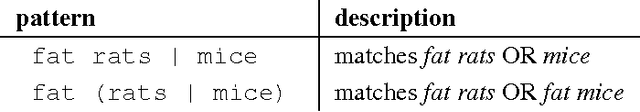

Abstract:This document describes the Odin framework, which is a domain-independent platform for developing rule-based event extraction models. Odin aims to be powerful (the rule language allows the modeling of complex syntactic structures) and robust (to recover from syntactic parsing errors, syntactic patterns can be freely mixed with surface, token-based patterns), while remaining simple (some domain grammars can be up and running in minutes), and fast (Odin processes over 100 sentences/second in a real-world domain with over 200 rules). Here we include a thorough definition of the Odin rule language, together with a description of the Odin API in the Scala language, which allows one to apply these rules to arbitrary texts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge