Marco A. Valenzuela-Escarcega
From Examples to Rules: Neural Guided Rule Synthesis for Information Extraction
Jan 16, 2022



Abstract:While deep learning approaches to information extraction have had many successes, they can be difficult to augment or maintain as needs shift. Rule-based methods, on the other hand, can be more easily modified. However, crafting rules requires expertise in linguistics and the domain of interest, making it infeasible for most users. Here we attempt to combine the advantages of these two directions while mitigating their drawbacks. We adapt recent advances from the adjacent field of program synthesis to information extraction, synthesizing rules from provided examples. We use a transformer-based architecture to guide an enumerative search, and show that this reduces the number of steps that need to be explored before a rule is found. Further, we show that without training the synthesis algorithm on the specific domain, our synthesized rules achieve state-of-the-art performance on the 1-shot scenario of a task that focuses on few-shot learning for relation classification, and competitive performance in the 5-shot scenario.
AutoMATES: Automated Model Assembly from Text, Equations, and Software
Jan 21, 2020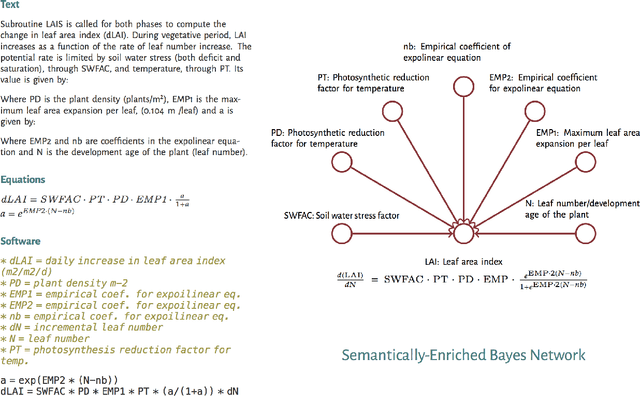
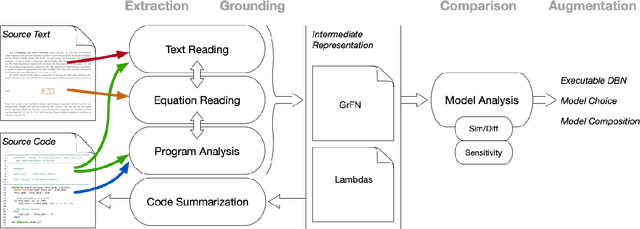
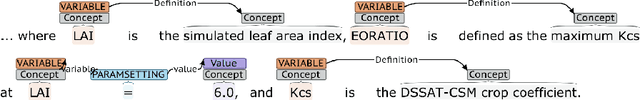
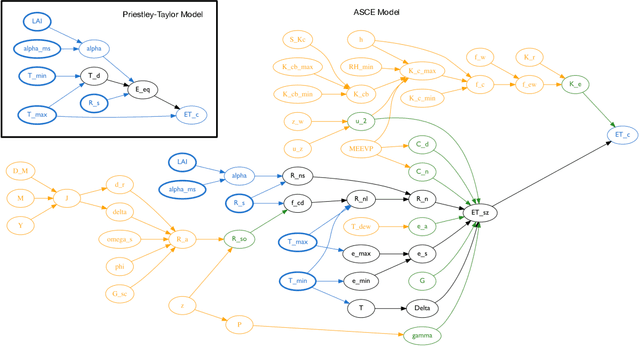
Abstract:Models of complicated systems can be represented in different ways - in scientific papers, they are represented using natural language text as well as equations. But to be of real use, they must also be implemented as software, thus making code a third form of representing models. We introduce the AutoMATES project, which aims to build semantically-rich unified representations of models from scientific code and publications to facilitate the integration of computational models from different domains and allow for modeling large, complicated systems that span multiple domains and levels of abstraction.
SnapToGrid: From Statistical to Interpretable Models for Biomedical Information Extraction
Jun 30, 2016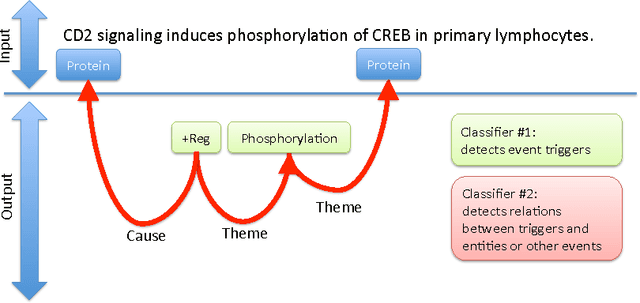
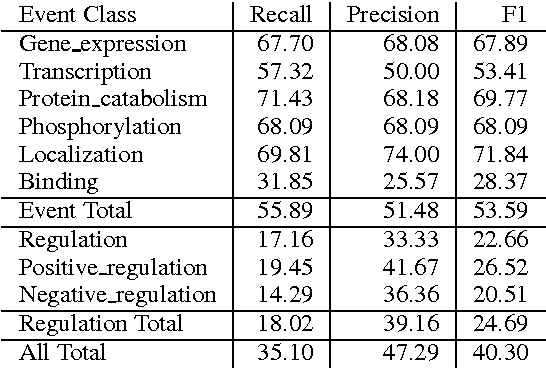
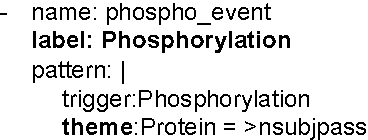
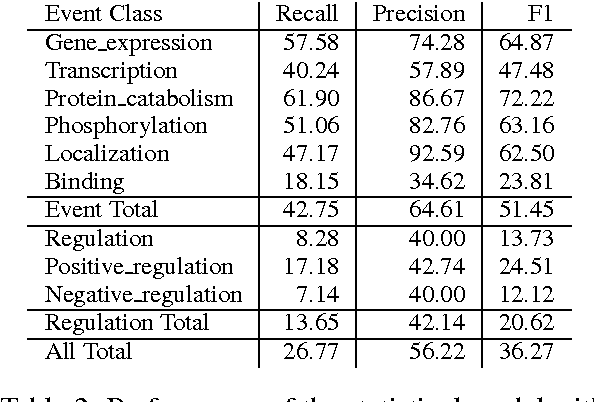
Abstract:We propose an approach for biomedical information extraction that marries the advantages of machine learning models, e.g., learning directly from data, with the benefits of rule-based approaches, e.g., interpretability. Our approach starts by training a feature-based statistical model, then converts this model to a rule-based variant by converting its features to rules, and "snapping to grid" the feature weights to discrete votes. In doing so, our proposal takes advantage of the large body of work in machine learning, but it produces an interpretable model, which can be directly edited by experts. We evaluate our approach on the BioNLP 2009 event extraction task. Our results show that there is a small performance penalty when converting the statistical model to rules, but the gain in interpretability compensates for that: with minimal effort, human experts improve this model to have similar performance to the statistical model that served as starting point.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge