Paul D. Hein
AutoMATES: Automated Model Assembly from Text, Equations, and Software
Jan 21, 2020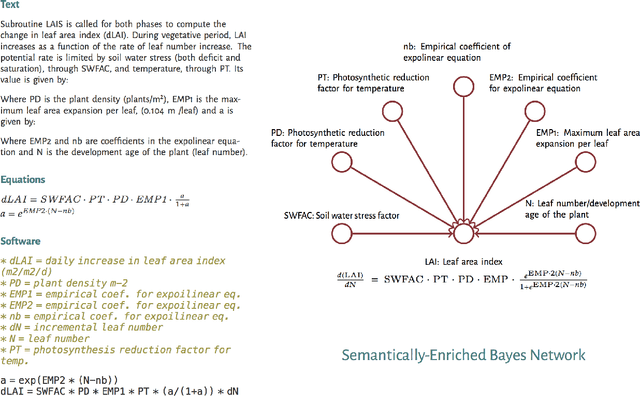
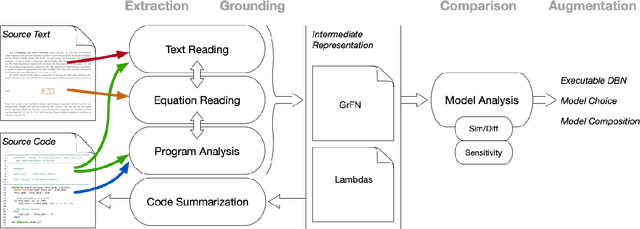
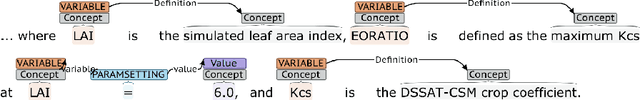
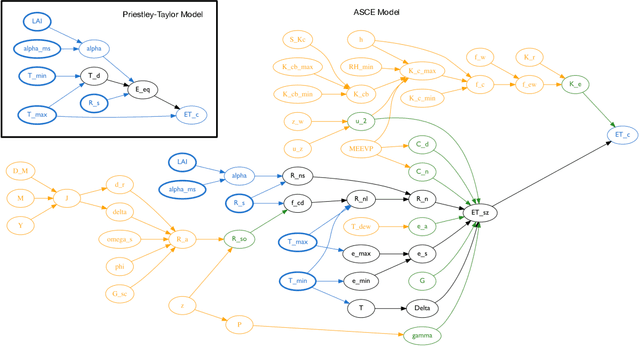
Abstract:Models of complicated systems can be represented in different ways - in scientific papers, they are represented using natural language text as well as equations. But to be of real use, they must also be implemented as software, thus making code a third form of representing models. We introduce the AutoMATES project, which aims to build semantically-rich unified representations of models from scientific code and publications to facilitate the integration of computational models from different domains and allow for modeling large, complicated systems that span multiple domains and levels of abstraction.
Meta-Learning Initializations for Image Segmentation
Dec 13, 2019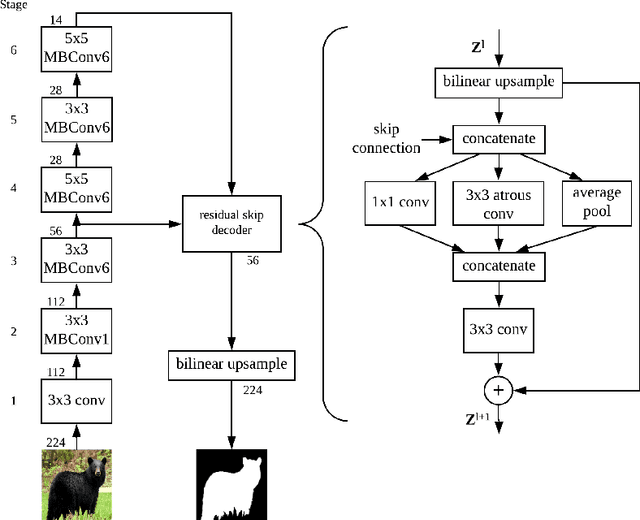
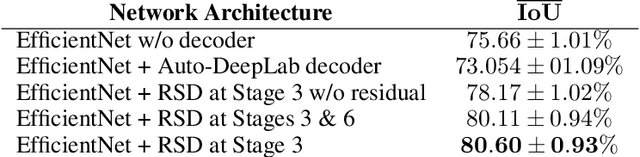
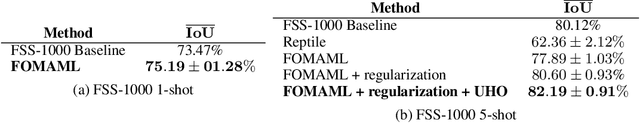
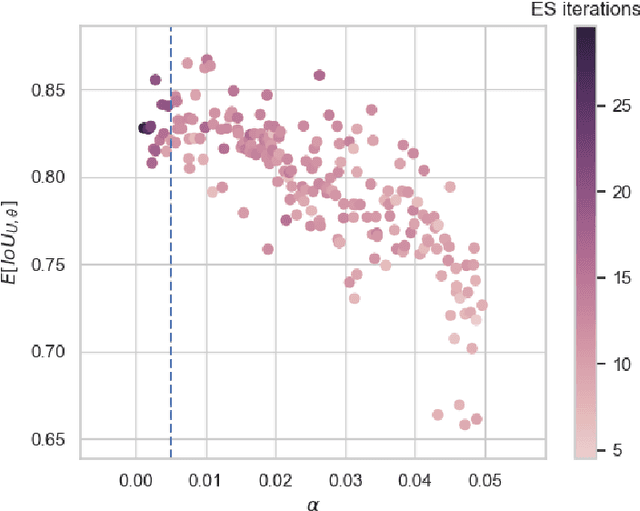
Abstract:While meta-learning approaches that utilize neural network representations have made progress in few-shot image classification, reinforcement learning, and, more recently, image semantic segmentation, the training algorithms and model architectures have become increasingly specialized to the few-shot domain. A natural question that arises is how to develop learning systems that scale from few-shot to many-shot settings while yielding competitive performance in both. One scalable potential approach that does not require ensembling many models nor the computational costs of relation networks, is to meta-learn an initialization. In this work, we study first-order meta-learning of initializations for deep neural networks that must produce dense, structured predictions given an arbitrary amount of training data for a new task. Our primary contributions include (1), an extension and experimental analysis of first-order model agnostic meta-learning algorithms (including FOMAML and Reptile) to image segmentation, (2) a novel neural network architecture built for parameter efficiency and fast learning which we call EfficientLab, (3) a formalization of the generalization error of meta-learning algorithms, which we leverage to decrease error on unseen tasks, and (4) a small benchmark dataset, FP-k, for the empirical study of how meta-learning systems perform in both few- and many-shot settings. We show that meta-learned initializations for image segmentation provide value for both canonical few-shot learning problems and larger datasets, outperforming ImageNet-trained initializations for up to 400 densely labeled examples. We find that our network, with an empirically estimated optimal update procedure, yields state of the art results on the FSS-1000 dataset while only requiring one forward pass through a single model at evaluation time.
Inter-sentence Relation Extraction for Associating Biological Context with Events in Biomedical Texts
Dec 14, 2018



Abstract:We present an analysis of the problem of identifying biological context and associating it with biochemical events in biomedical texts. This constitutes a non-trivial, inter-sentential relation extraction task. We focus on biological context as descriptions of the species, tissue type and cell type that are associated with biochemical events. We describe the properties of an annotated corpus of context-event relations and present and evaluate several classifiers for context-event association trained on syntactic, distance and frequency features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge