Enrique Noriega-Atala
Variable Extraction for Model Recovery in Scientific Literature
Nov 21, 2024



Abstract:The global output of academic publications exceeds 5 million articles per year, making it difficult for humans to keep up with even a tiny fraction of scientific output. We need methods to navigate and interpret the artifacts -- texts, graphs, charts, code, models, and datasets -- that make up the literature. This paper evaluates various methods for extracting mathematical model variables from epidemiological studies, such as ``infection rate ($\alpha$),'' ``recovery rate ($\gamma$),'' and ``mortality rate ($\mu$).'' Variable extraction appears to be a basic task, but plays a pivotal role in recovering models from scientific literature. Once extracted, we can use these variables for automatic mathematical modeling, simulation, and replication of published results. We introduce a benchmark dataset comprising manually-annotated variable descriptions and variable values extracted from scientific papers. Based on this dataset, we present several baseline methods for variable extraction based on Large Language Models (LLMs) and rule-based information extraction systems. Our analysis shows that LLM-based solutions perform the best. Despite the incremental benefits of combining rule-based extraction outputs with LLMs, the leap in performance attributed to the transfer-learning and instruction-tuning capabilities of LLMs themselves is far more significant. This investigation demonstrates the potential of LLMs to enhance automatic comprehension of scientific artifacts and for automatic model recovery and simulation.
When and Where Did it Happen? An Encoder-Decoder Model to Identify Scenario Context
Oct 10, 2024



Abstract:We introduce a neural architecture finetuned for the task of scenario context generation: The relevant location and time of an event or entity mentioned in text. Contextualizing information extraction helps to scope the validity of automated finings when aggregating them as knowledge graphs. Our approach uses a high-quality curated dataset of time and location annotations in a corpus of epidemiology papers to train an encoder-decoder architecture. We also explored the use of data augmentation techniques during training. Our findings suggest that a relatively small fine-tuned encoder-decoder model performs better than out-of-the-box LLMs and semantic role labeling parsers to accurate predict the relevant scenario information of a particular entity or event.
Learning Open Domain Multi-hop Search Using Reinforcement Learning
May 30, 2022
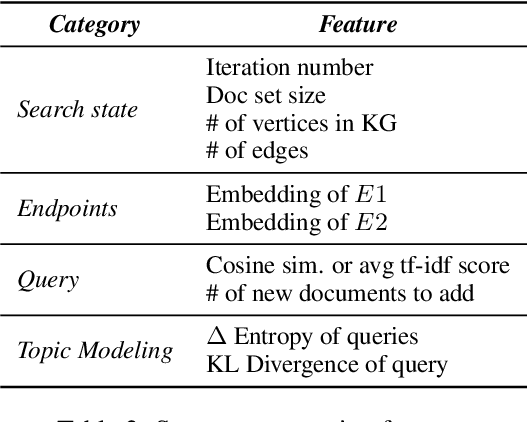
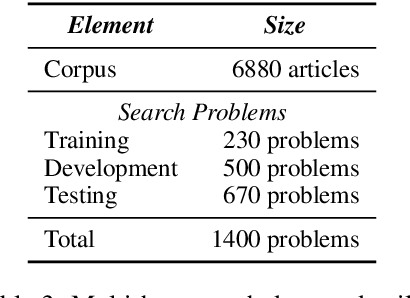
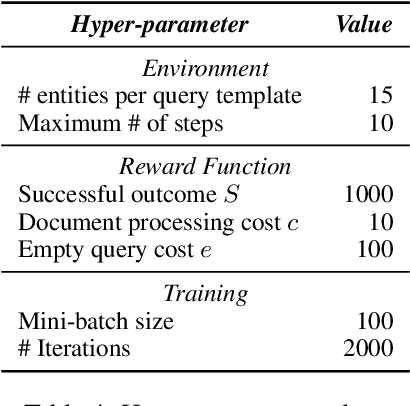
Abstract:We propose a method to teach an automated agent to learn how to search for multi-hop paths of relations between entities in an open domain. The method learns a policy for directing existing information retrieval and machine reading resources to focus on relevant regions of a corpus. The approach formulates the learning problem as a Markov decision process with a state representation that encodes the dynamics of the search process and a reward structure that minimizes the number of documents that must be processed while still finding multi-hop paths. We implement the method in an actor-critic reinforcement learning algorithm and evaluate it on a dataset of search problems derived from a subset of English Wikipedia. The algorithm finds a family of policies that succeeds in extracting the desired information while processing fewer documents compared to several baseline heuristic algorithms.
Neural Architectures for Biological Inter-Sentence Relation Extraction
Dec 17, 2021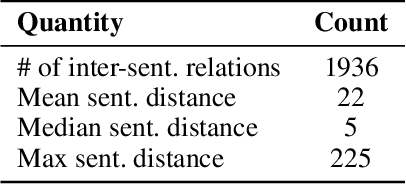

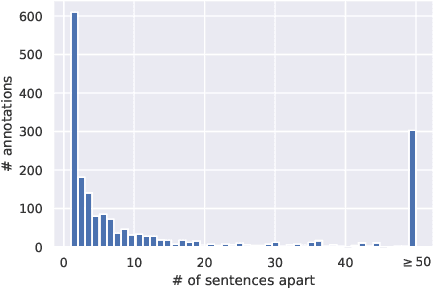

Abstract:We introduce a family of deep-learning architectures for inter-sentence relation extraction, i.e., relations where the participants are not necessarily in the same sentence. We apply these architectures to an important use case in the biomedical domain: assigning biological context to biochemical events. In this work, biological context is defined as the type of biological system within which the biochemical event is observed. The neural architectures encode and aggregate multiple occurrences of the same candidate context mentions to determine whether it is the correct context for a particular event mention. We propose two broad types of architectures: the first type aggregates multiple instances that correspond to the same candidate context with respect to event mention before emitting a classification; the second type independently classifies each instance and uses the results to vote for the final class, akin to an ensemble approach. Our experiments show that the proposed neural classifiers are competitive and some achieve better performance than previous state of the art traditional machine learning methods without the need for feature engineering. Our analysis shows that the neural methods particularly improve precision compared to traditional machine learning classifiers and also demonstrates how the difficulty of inter-sentence relation extraction increases as the distance between the event and context mentions increase.
Inter-sentence Relation Extraction for Associating Biological Context with Events in Biomedical Texts
Dec 14, 2018



Abstract:We present an analysis of the problem of identifying biological context and associating it with biochemical events in biomedical texts. This constitutes a non-trivial, inter-sentential relation extraction task. We focus on biological context as descriptions of the species, tissue type and cell type that are associated with biochemical events. We describe the properties of an annotated corpus of context-event relations and present and evaluate several classifiers for context-event association trained on syntactic, distance and frequency features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge