SnapToGrid: From Statistical to Interpretable Models for Biomedical Information Extraction
Paper and Code
Jun 30, 2016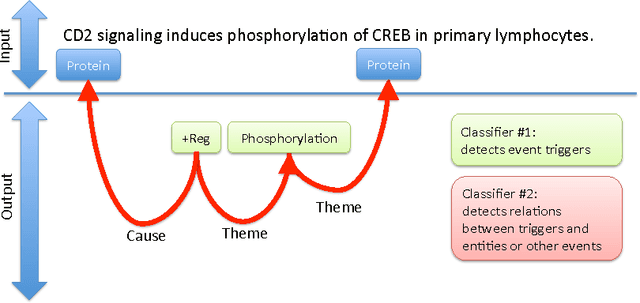
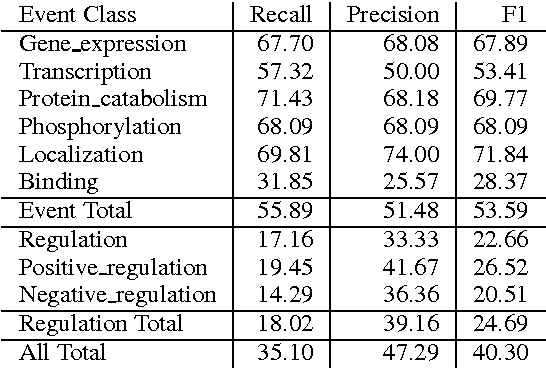
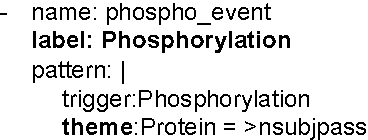
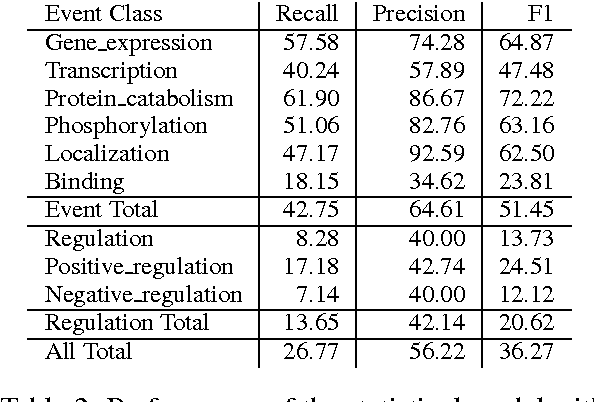
We propose an approach for biomedical information extraction that marries the advantages of machine learning models, e.g., learning directly from data, with the benefits of rule-based approaches, e.g., interpretability. Our approach starts by training a feature-based statistical model, then converts this model to a rule-based variant by converting its features to rules, and "snapping to grid" the feature weights to discrete votes. In doing so, our proposal takes advantage of the large body of work in machine learning, but it produces an interpretable model, which can be directly edited by experts. We evaluate our approach on the BioNLP 2009 event extraction task. Our results show that there is a small performance penalty when converting the statistical model to rules, but the gain in interpretability compensates for that: with minimal effort, human experts improve this model to have similar performance to the statistical model that served as starting point.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge